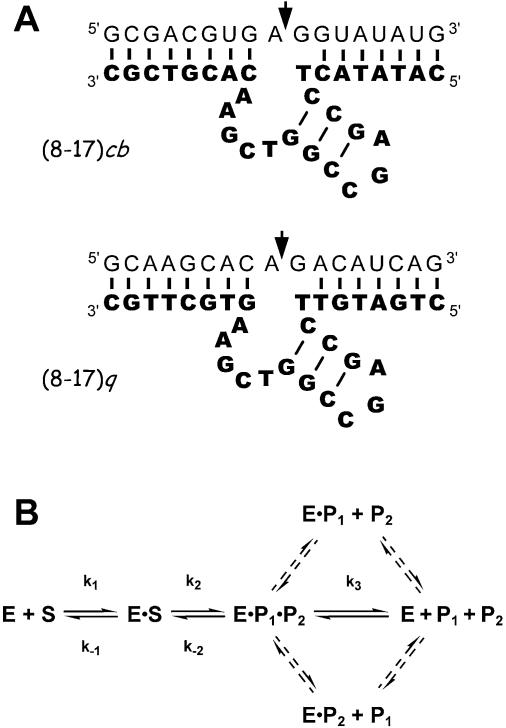
Figure 1.
(A) Primary and secondary structures of the 8-17 constructs used in this study. The DNA enzymes are shown in bold letters and their RNA substrates are shown in normal letters. The arrows indicate the sites of cleavage. (B) Kinetic scheme for the RNA cleavage reaction carried out by the 8-17 deoxyribozyme. The DNAzyme (E) binds to its substrate (S), forming a productive bimolecular complex (E·S). Subsequently, S is cleaved to yield two products: P1 (containing a cyclic 2′,3′-phosphate) and P2 (containing a free 3′-hydroxyl) (4). P1 and P2 can dissociate with different rates and in different orders (dashed arrows); in general, however, only release of the product that dissociates most slowly affects multiple-turnover catalysis. This rate is represented here by k3.