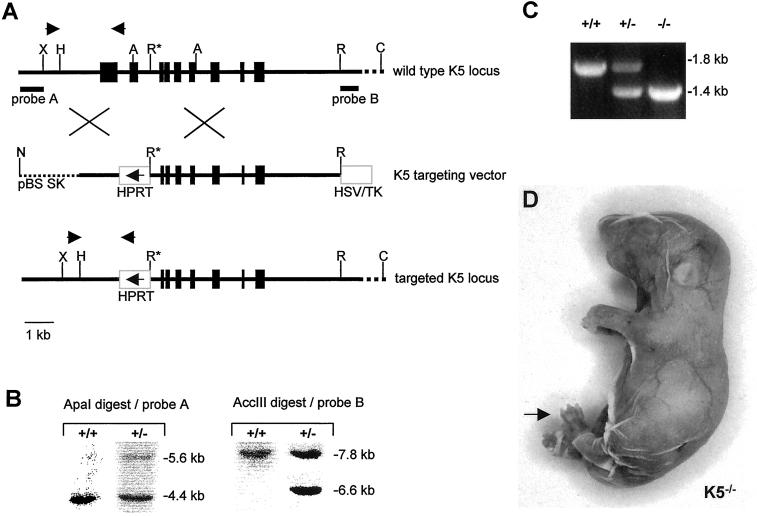
Figure 1.
Targeting strategy and confirmation of gene targeting event. (A) Map of the K5 gene locus, the targeting vector, and the recombinant K5 locus. The core promoter and the first two exons of the K5 gene up to the 5′ EcoRI (R*) site were replaced by the HPRT minigene. The arrow in the HPRT minigene indicates the direction of transcription. In addition, an HSV/TK minigene was inserted as a negative selectable marker. The NotI restriction site was used to linearize the vector for transfection. Probes A and B mark the position of the 5′ and 3′ probe used in Southern blotting. Arrows above the K5 gene locus and the recombinant allele indicate primer positions for PCR-based genotyping. Letters indicate restriction sites: A, ApaI; C, AccIII; H, HindIII; N, NotI; R, EcoRI; X, XbaI. (B) Example of Southern blot analysis of ES cells. To confirm the correct targeting event, genomic DNA was digested with ApaI for detection with the 5′ probe, which led to a 5.6-kb band for the targeted allele and a 4.4-kb band for the wild-type allele. For detection with the 3′ probe, an AccIII digest was performed resulting in a 6.6-kb fragment for the targeted allele and a 7.8-kb fragment for the wild-type allele. (C) Identification of genotypes by PCR. Primers designed to identify wild-type and mutant alleles (A) were used for genotyping of the litters. The wild-type allele resulted in a product of ∼1.8-kb size, the targeted allele in a 1.4-kb product. (D) Neonatal homozygous K5−/− mouse. The fragile epidermis almost completely lost contact with the dermis after the mechanical stress of birth. Paws were sometimes denuded (arrow). K5−/− animals died within 1 h after birth.