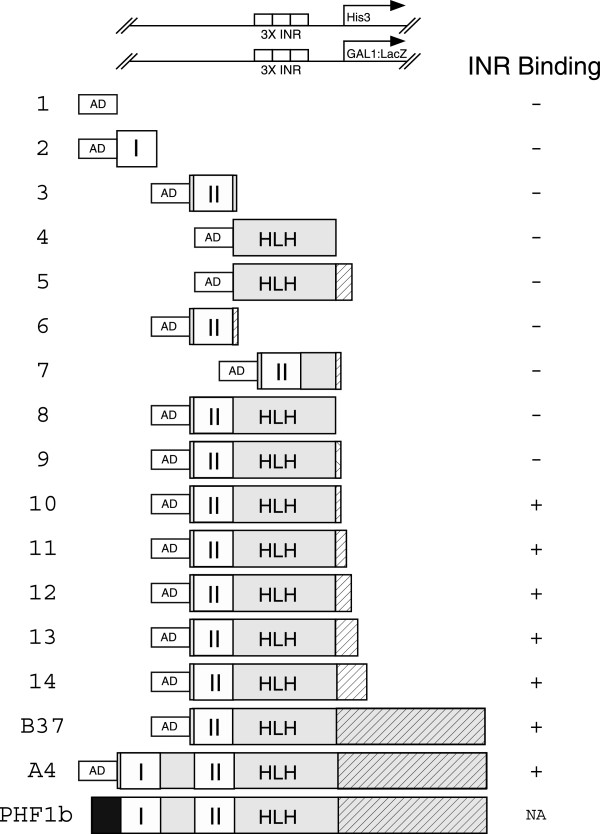
Figure 2.
Mapping sequences that are required for PHF1b DNA recognition. Depicted is the yeast strain with chromosomally integrated reporters carrying three tandem β1 initiator sites that was used for one-hybrid assays. Candidates identified by one-hybrid assays were used to construct a number of 5′ and 3′ sequential deletions of the PHF1b gene to define the minimal β1-INR binding domain. The GAL4 AD was fused to all of the PHF1b fragments and tested for its ability to activate three tandem β1-INR sites linked to either His3 or LacZ. Full length PHF1b is shown for comparison to relate relative sizes of AD fused PHF1b fragments (#2-14 with the exception of #9, which is a derivative of PHF1a) and one-hybrid candidates B37 and A4. The DNA binding activities both positive (+) and negative (−) are as indicated. The criterion for DNA binding is measured by the ability of the constructs to activate both reporter genes (His3 and LacZ ) at levels comparable to B37 and A4 isolates. His3 expression was measured by the survival of the yeast colonies that could grow on plates that contained 10 mM 3-aminotriazole and β-galactosidase activity from the second reporter, measured by β-galactosidase assays. The results from both assays were the same and are indicated by one column to the right.