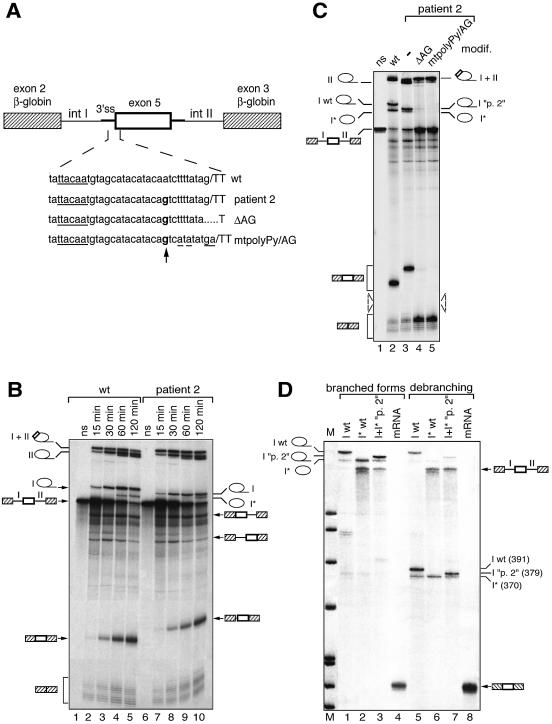
Figure 5.
In vitro splicing assays of wild-type and patient 2 (IVS4 –11 A→G) transcripts. (A) Chimeric constructs used in in vitro splicing assays are as in Figure 2B. Chimeric introns upstream and downstream of exon 5, arbitrarily designated introns I and II, are 384 and 653 nt long, respectively. The site of the mutation is indicated in bold and the consensus defining the branch site is underlined. The mutations altering the natural 3′ AG of patient 2 are also indicated. (B) Kinetic analysis of splicing. Splicing reactions were run for 15, 30, 60 and 120 min and RNA products were analysed by PAGE. Selected splicing products are indicated along the gel panel. (C) Comparison of splicing products generated after 120 min of splicing from the wild-type transcript (wt) and patient 2 transcripts carrying (lanes 4–5) or not (lane 3) mutations in the natural 3′ AG. (D) Comparative analysis of lariat intron I molecules produced from wild-type (wt) and mutant (“p. 2”) transcripts. Isolated putative intron I wt (I wt), shortened intron I wt (I* wt) as well as intact and shortened forms of intron I “p. 2” together (I+I* “p. 2”) were analysed directly (lanes 1–3) or after debranching (lanes 5–7). A linear three-exon mRNA was also analysed as a control before (lane 4) or after (lane 8) debranching. The size of the debranched RNAs was determined using DNA size markers (pBR322 cut by MspI), run in parallel.