Figure 7.
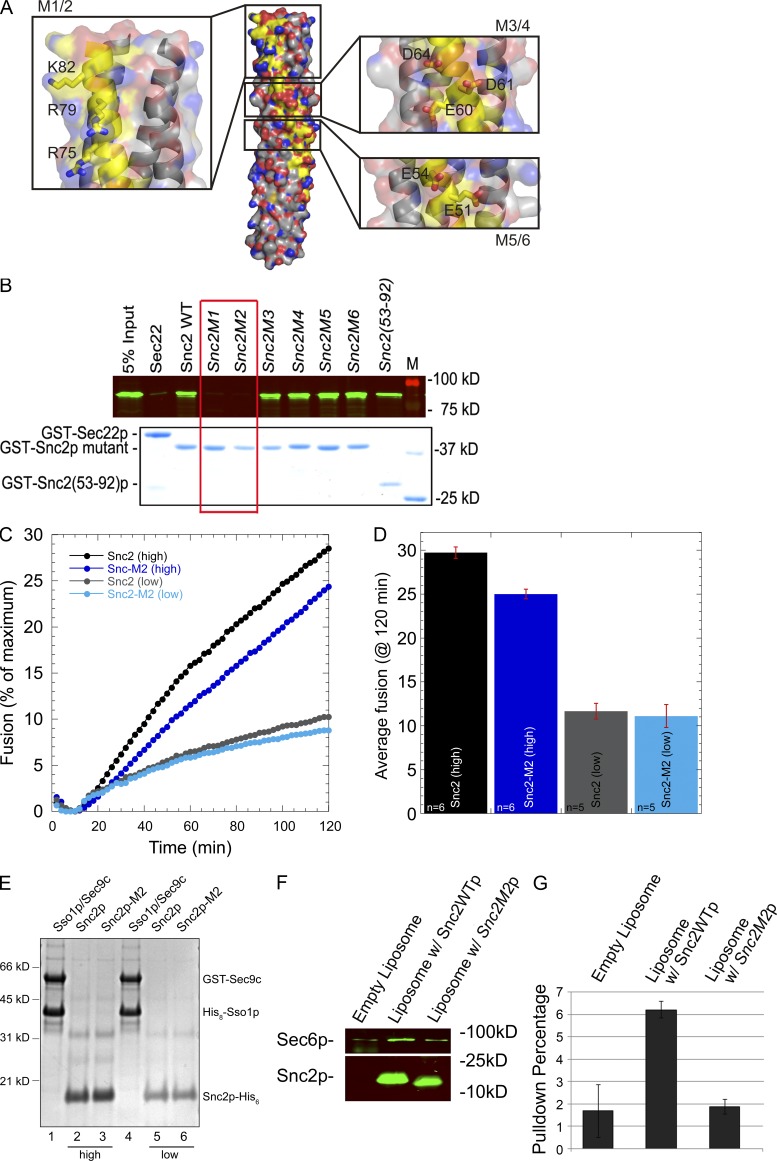
A positively charged patch at the end of the Snc2p SNARE domain is needed for Sec6p binding, but not liposome fusion. (A) The structure of the Snc2p–Ssop–Sec9p SNARE complex is shown with the Snc2p backbone highlighted in yellow and the three charged patches on the surface of Snc2p expanded in insets with side chains shown. Images were generated by PyMol using coordinates of 3B5N from the PDB provided by Strop et al. (2008). Snc1p in the original structure was replaced with Snc2p. (B) Binding of Sec6p to Snc2p mutants. His-tagged Sec6p (80 nM final concentration) was mixed with 50 nM (final concentration) of the various Snc2p mutants or with GST-Sec22p as a control. Both M1 (R75A R79A K82A) and M2 (R75E R79E K82E) show a loss of Sec6p binding activity, whereas M3 (E60A D61A D64A), M4 (E60K D61R D64R), M5 (E51A E54A), and M6 (E51R E54R) bind normally. (C) Kinetic fusion assay comparing different donor v-SNARE liposomes containing either wild-type Snc2p or Snc2p-M2 mutant. t-SNARE liposomes containing GST-Sec9c and His6-Sso1p (45 µl) were mixed with 5 µl v-SNARE liposomes and NBD fluorescence was monitored in a fluorescent plate reader for 2 h. Fusion with t-SNAREs (∼460 pmol of t-SNARE complex proteins, ∼55.8 nmol lipid) and wild-type Snc2p (high, ∼136 pmol protein, 4.5 nmol lipid) are shown in black circles, whereas fusion with Snc2p-M2 (high, ∼192 pmol protein, 4.6 nmol lipid) are shown in dark blue. Similarly, t-SNARE liposomes with less total protein (∼200 pmol of t-SNARE complex proteins, ∼28.3 nmol lipid) were fused with wild-type Snc2 liposomes (low, 21 pmol protein, 2.2 nmol lipid, gray circles) or Snc2p-M2 liposomes (low, ∼15 pmol protein, 1.3 nmol lipid, light blue circles). Representative traces are shown. (D) Average endpoint fusion. The black and dark blue bar graphs represent fusion with high concentrations of the v-SNARE protein (wild-type and mutant) in the liposomes, whereas gray and light blue bar graphs represent fusion with low concentrations of the v-SNARE proteins (wild type and mutant) in the donor liposomes. The histograms show mean fusion at 120 min and the error bars represent SEM. The number of replicates (n) is shown at the base of the histogram. (E) Coomassie blue–stained gel of proteoliposomes on a 10% Bis-Tris NuPAGE Novex gel (Invitrogen). Lanes 1–4 contain 5 µl of proteoliposomes, whereas lanes 5 and 6 (low concentration v-SNAREs) contain 10 µl of proteoliposomes. (F) Sec6p binds to Snc2p reconstituted into liposomes. Empty liposome (0.01 mM lipid) or liposomes containing 0.4 µM Snc2p or Snc2p-M2 were incubated with 0.025 µM His-Sec6 (final concentration) at 4°C for 2 h in 1 ml binding buffer (25 mM Hepes-KOH and 200 mM KCl, pH 7.4, with 0.5 µg/µl BSA). Liposomes were pelleted at 300,000 g for 20 min in an ultracentrifuge (Optima TLX; Beckman Coulter) with a TLA 120.2 rotor. The pellet was washed with 1 ml binding buffer and re-pelleted. The pellet was resuspended in 80 µl sample buffer. 10 µl of each sample was resolved by SDS-PAGE and analyzed by Western blot using anti-His (top) and anti-Snc (bottom) antibody. (G) Quantitation of Sec6p binding.