Figure 2.
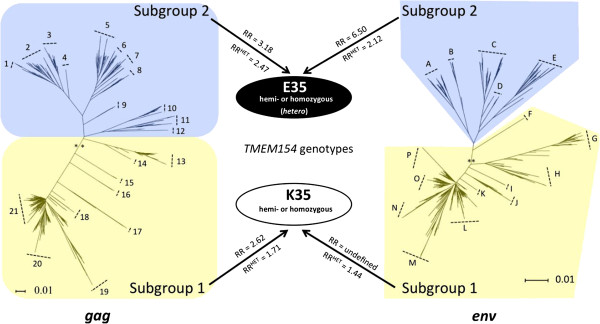
Identification of SRLV gag and env subgroups that associate with TMEM154 E35K genotypes. The gag and env Neighbor-Joining trees represent all 655 gag and 449 env sequences produced in this study, respectively. Numbers and letters represent phylotype designations in the gag and env trees, respectively. Double asterisks show the mid-point root site of the trees. Scale bars represent substitutions per site. Yellow and blue shaded regions represent subgroups 1 and 2, respectively, for both trees. Arrows show the relative risks of TMEM154 E35 and K35 sheep infection by SRLV subgroups. RRHETs were derived with grouped hemi-, hetero-, and homozygous TMEM154 E35 sheep; and grouped hemi-and homozygous K35 sheep. RRs were derived with hemi and homozygous groups of TMEM154 E35 sheep and K35 sheep, respectively. All hemizygous genotypes consisted of one copy of haplotype 4, which contained a deletion mutation and did not code for a predicted full-length biologically active isomer.
