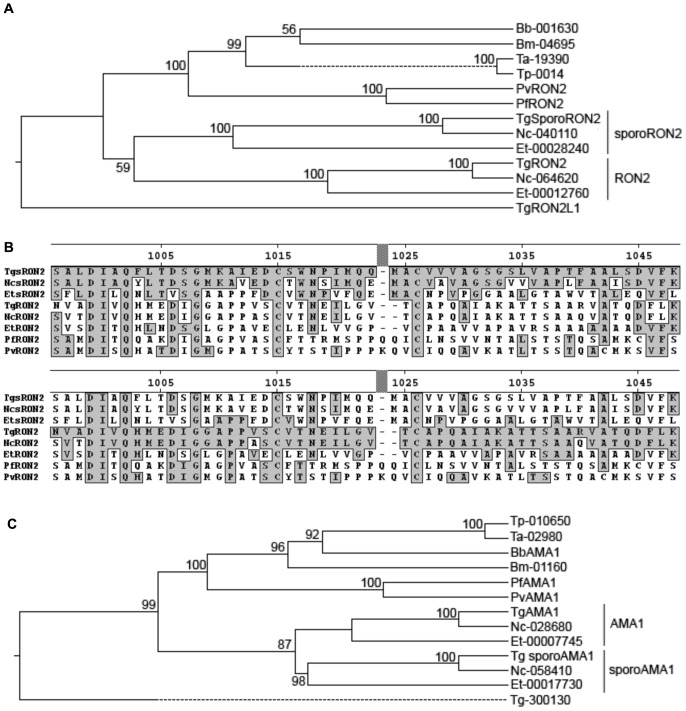
Figure 1. T. gondii sporoRON2 and sporoAMA1 are conserved in other Apicomplexans and are distinct from generic RON2 and generic AMA1.
A. The Toxoplasma sporoRON2 polypeptide sequences were aligned with their respective homologues in Eimeria tenella, Neospora caninum, Plasmodium spp. (P. falciparum and P. vivax), Babesia spp. (B.bovis and B.microti), and Theileria spp. (T. annulata and T. parva) using ClustalW (as part of MegAlign software (Lasergene)) and an anchored phylogenetic tree was generated using the standard algorithm. The clusters including generic and sporozoite-specific versions of each protein are so-labeled. Bootstrapping analysis was performed to determine confidence intervals (1000 trials). B. Alignment of domain 3 (D3) of the indicated RON2 homologues was performed in ClustalW. Residues identical to that of T. gondii sporoRON2 are indicated with shading on the upper panel, while residues identical to those of T. gondii generic RON2 are boxed on the lower panel. Numbers indicate amino acid position of T. gondii sporoRON2 from the starting Methionine. C. As for (A) except using the predicted sporoAMA1 polypeptide sequences.