Figure 1.
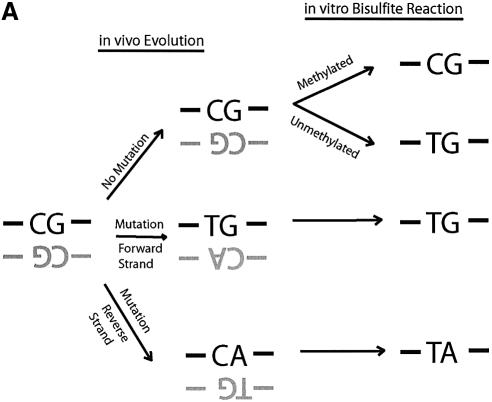
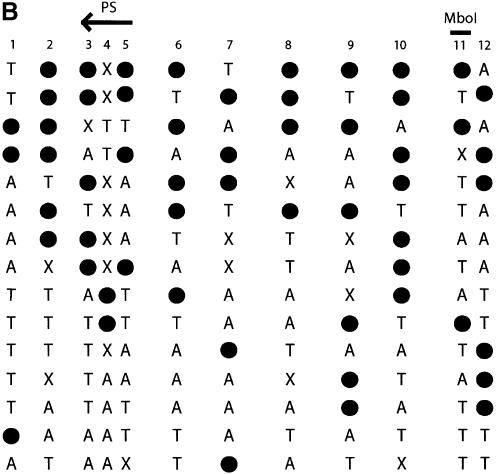
Direct DNA sequencing of bisulfite repetitive element PCR of Alu elements. (A) Schematic of possible fates of Alu element CpG methylation sites. Due to the mutation of CpG sites via spontaneous deamination of 5-methylcytosine to T during evolution, a CpG site can be changed into a TpG or CpA dinucleotide. Neither TpG nor CpA are targets for methylation. Following bisulfite treatment, a methylated CpG will remain CpG. However, an unmethylated CpG will give rise to TpG, which is indistinguishable from a deamination mutation of the forward strand. Thus, three possibilities arise for our sequencing data: CpG that represents a methylated CpG site, TpG that represents either an unmethylated CpG site or a mutation of the forward strand, and TpA that represents a mutation of the reverse strand followed by conversion of the unmethylated C to T by bisulfite. (B) Sequencing data of bisulfite repetitive element PCR of Alu elements. Genomic DNA was isolated from peripheral human blood and bisulfite treated. Alu element PCR was performed and the PCR product was cloned, and 15 clones were sequenced. There were 12 potential CpG methylation sites per clone for a total of 180 potential methylation sites sequenced. Black circles represent methylated CpG sites (66/180 = 36.7%). ‘T’ represents TpG sites that were either unmethylated or mutated (53/180 = 24.4%). ‘A’ represents TpA sites that were mutated (41/180 = 22.8%). ‘X’ represents other mutations (20/180 = 11.1%). The CpG sites used for pyrosequencing (‘PS’) and COBRA (‘MboI’) are indicated.