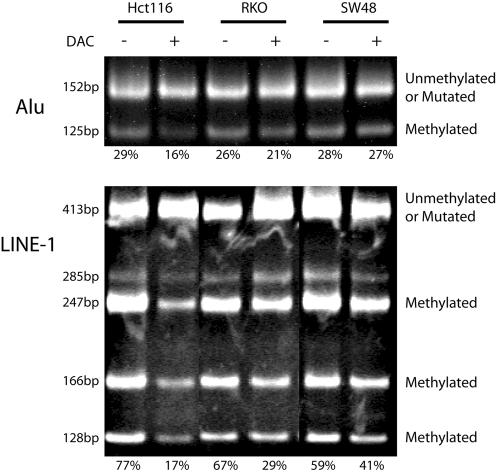
Figure 3.
Quantitation of DNA methylation in cell lines treated with DAC using the bisulfite repetitive element PCR technique. Hct116, RKO and SW48 cell lines were treated with DAC. Genomic DNA was isolated from DAC-treated (+) and -untreated (–) controls. The genomic DNA was treated with sodium bisulfite and a non-specific PCR was performed, which amplified a pool of Alu or LINE-1 repetitive elements. The PCR product was then digested with MboI (Alu) or HinfI (LINE-1), which only cuts repetitive elements that were originally methylated. The Alu PCR assays a single methylation site and therefore the MboI digestion will cut the PCR product from 152 to 125 and 27 bp (data not shown). The LINE-1 PCR assays the methylation of two sites and therefore HinfI can generate five possible digestion products of 285, 247, 166, 128 and 38 bp (data not shown). The digested PCR product was separated by polyacrylamide gel electrophoresis and stained with ethidium bromide. The lower cut bands represent methylated repetitive elements. The upper band represents unmethylated repetitive elements or repetitive elements in which the restriction site has been mutated. The PCR bands were quantitated and the amount of methylation is shown below each gel lane. Similar results were obtained for both the Alu element and LINE-1 assays.