Table I. Proteins identified via MS from leaves of different NaCl-treated T. halophila.
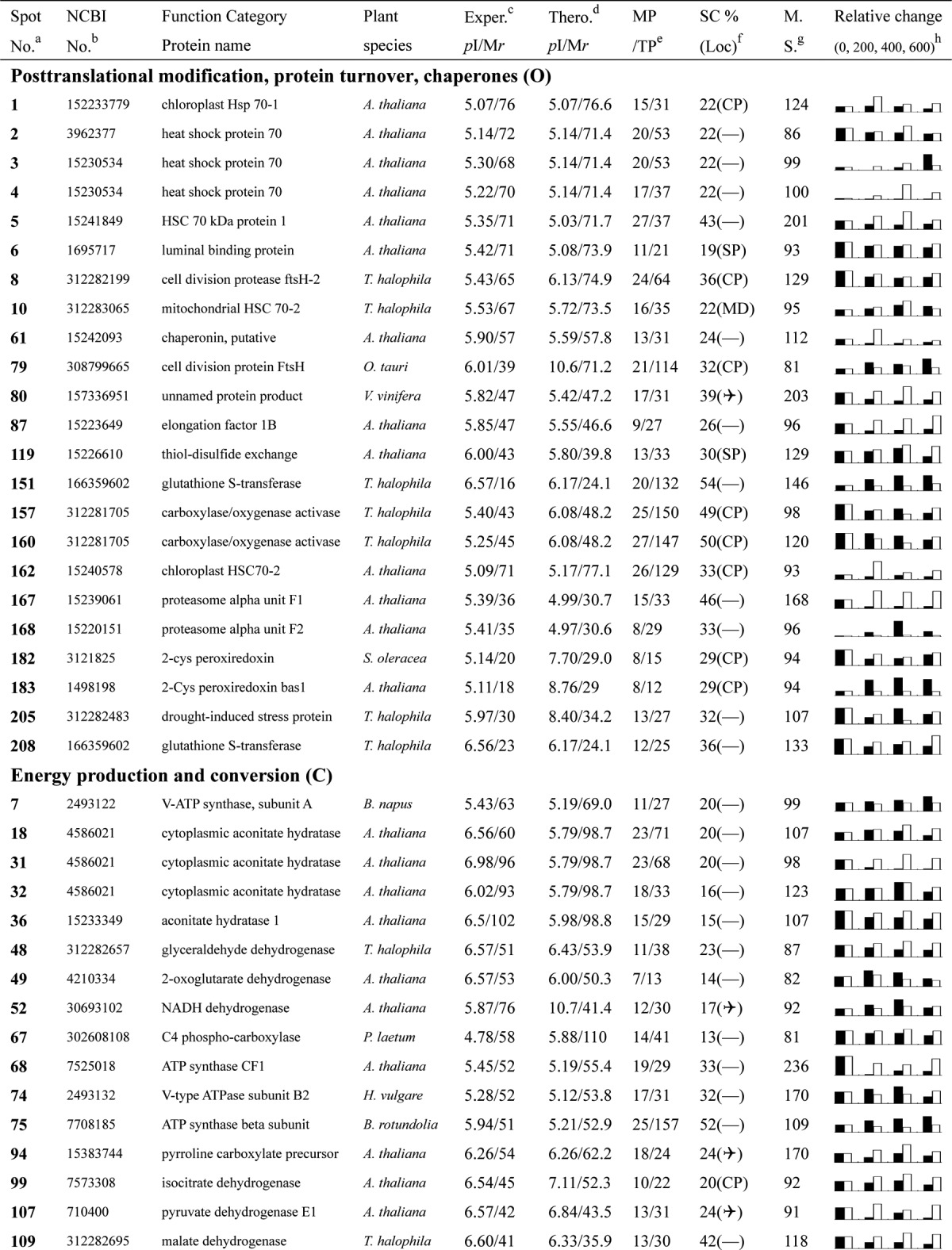
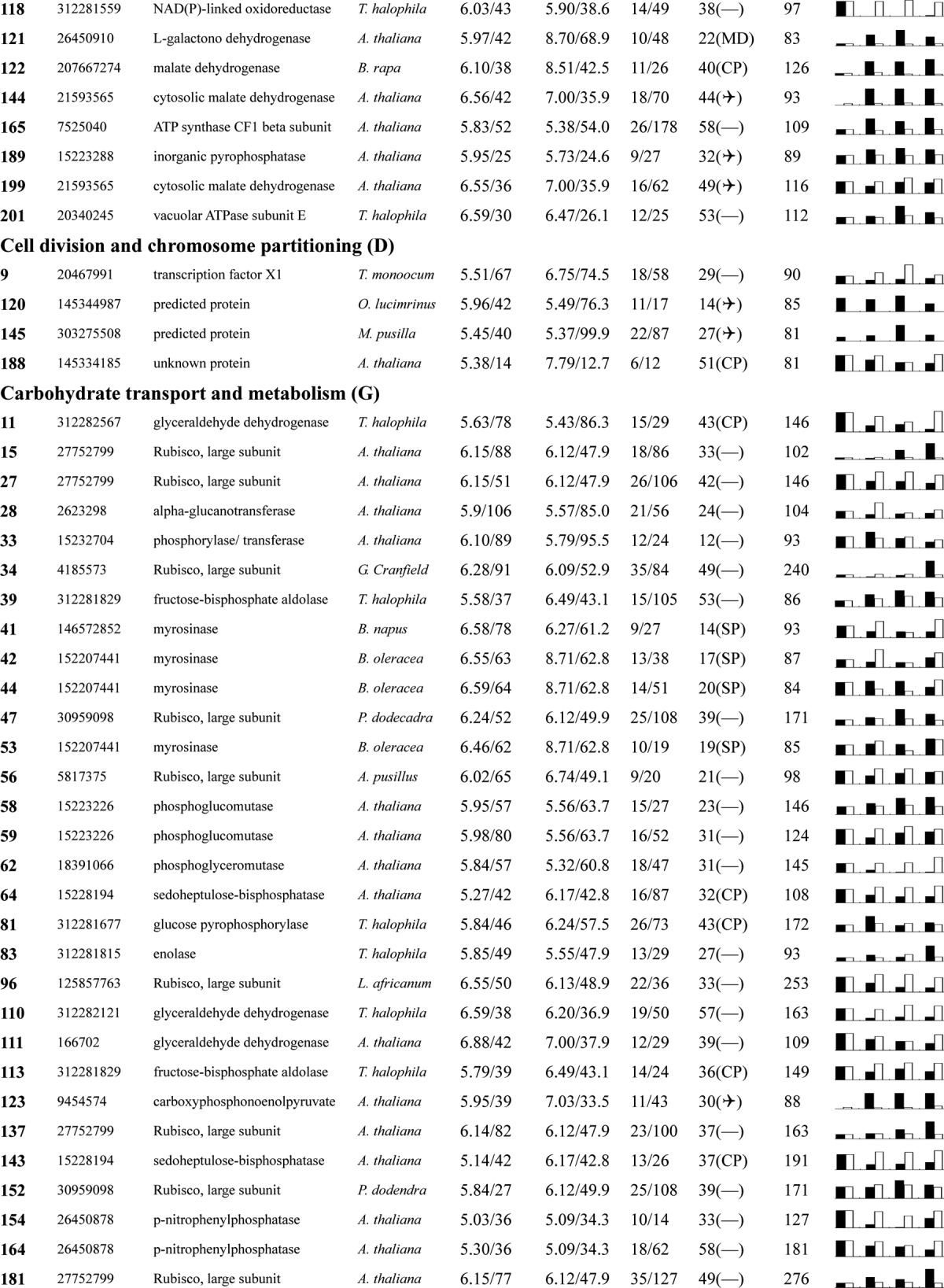
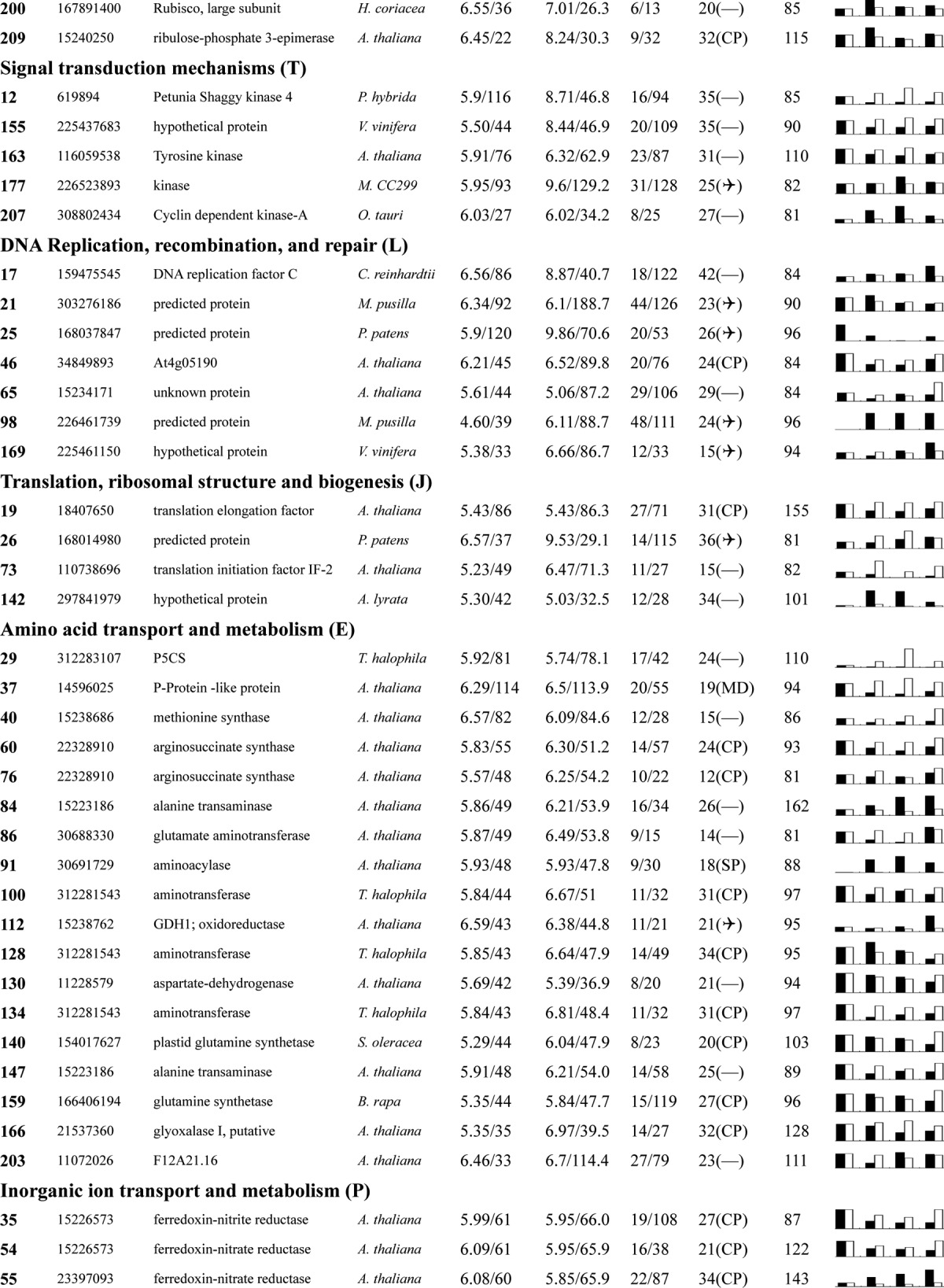
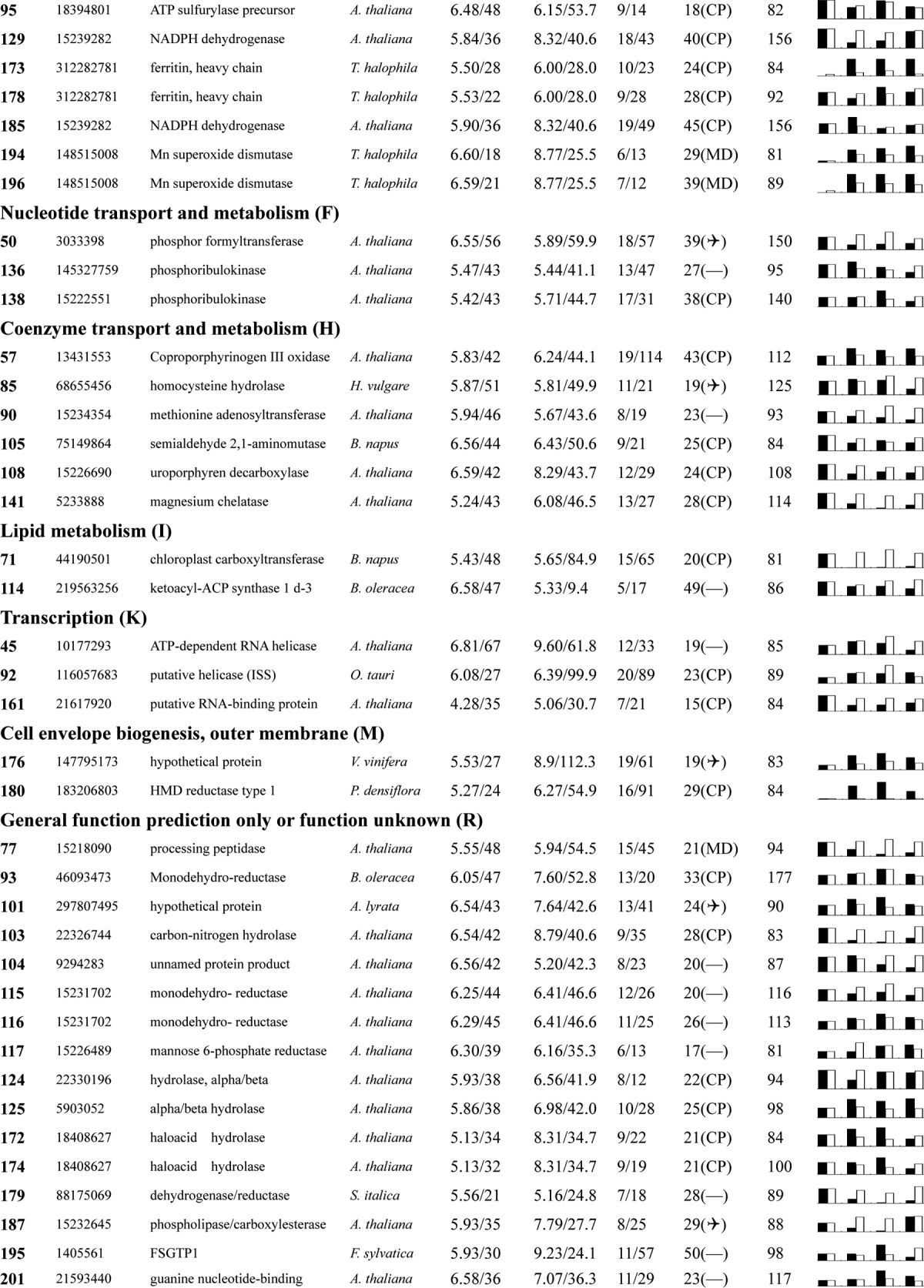
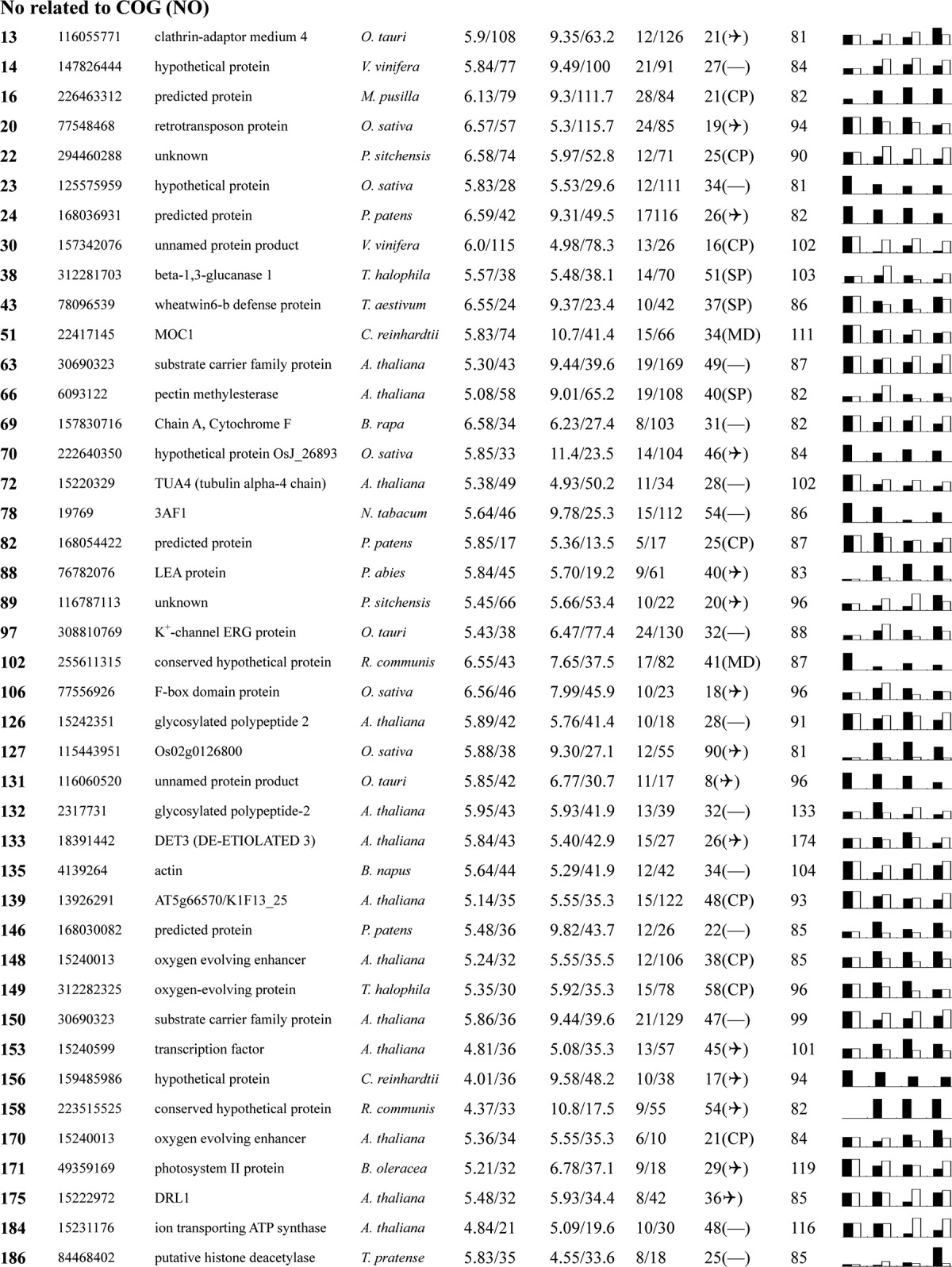
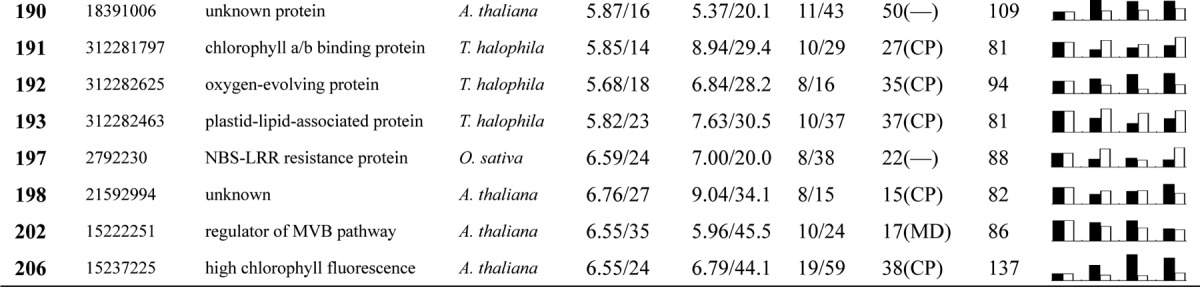
a Assigned spot number as indicated in Fig. 4.
b Database accession numbers according to NCBInr.
c Experimental mass (kDa) and pI of identified proteins were retrieved from the protein database. Experimental values were produced by ImageMaster.
d Theoretical mass (kDa) and pI of identified proteins were retrieved from the protein database.
e Number of the matched peptides (MP) with peptide mass fingerprinting and the total searched peptides (TP).
f The amino acid sequence coverage (SC) for the identified proteins and the protein location (Loc) predicted by TargetP.
g The Mascot searched score (M.S.) against NCBInr.
h Mean values of relative protein amounts on 2-DE gels (black bars) and mRNA changed patterns (white bars) in microarrays from the 0, 200, 400, and 600 mm NaCl treatments.
