Figure 1.
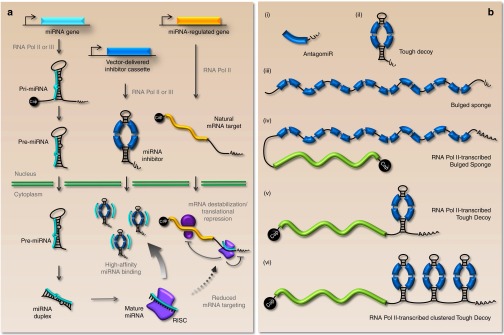
A schematic view of the anti-miR function of vector-encoded miRNA inhibitors. (a) Overview of miRNA biogenesis, miRNA regulation of protein-coding mRNAs, and miRNA inhibition by vector-encoded inhibitors. Long primary miRNA transcripts are transcribed from the genome and sequentially processed and transported to the cytoplasm, where they exert the posttranscriptional repression of target genes via the RNA-induced silencing complex (RISC). A miRNA inhibitor carrying miRNA binding sites (here represented by a Tough Decoy-type inhibitor) is intracellularly produced and interferes with natural miRNA regulation by attracting miRNAs by the exposure of high-affinity miRNA binding sites. (b) Schematic representation of commonly used inhibitor designs. (i) An antagomir consisting of a single miRNA binding site (shown in blue) followed by a short stretch of uridines originating from the RNA Pol III termination signal. (ii) An RNA Pol III-transcribed Tough Decoy consists of a ~60-bp long hairpin-shaped RNA carrying an internal loop exposing two miRNA binding sites each with a central bulge for prevention of fast endonucleolytic turnover of the transcript upon miRNA binding. The stretch of terminal uridines from the RNA Pol III termination signal provides the necessary 3' overhang for Exportin-5 recognition (iii) An RNA Pol III-transcribed Bulged Sponge design is generally expressed as a nonstructured RNA harboring 4–12 consecutive miRNA binding sites (eight are shown) with central bulges. (iv and v) RNA Pol II-transcribed variants of the Bulged Sponge and Tough Decoy resemble the Pol III-transcribed inhibitors, but are capped and polyadenylated and may be fused to a protein-coding sequence (shown in green). (vi) The Pol II-transcribed clustered Tough Decoy carrying three consecutive Tough Decoys with a total of six miRNA binding sites.
