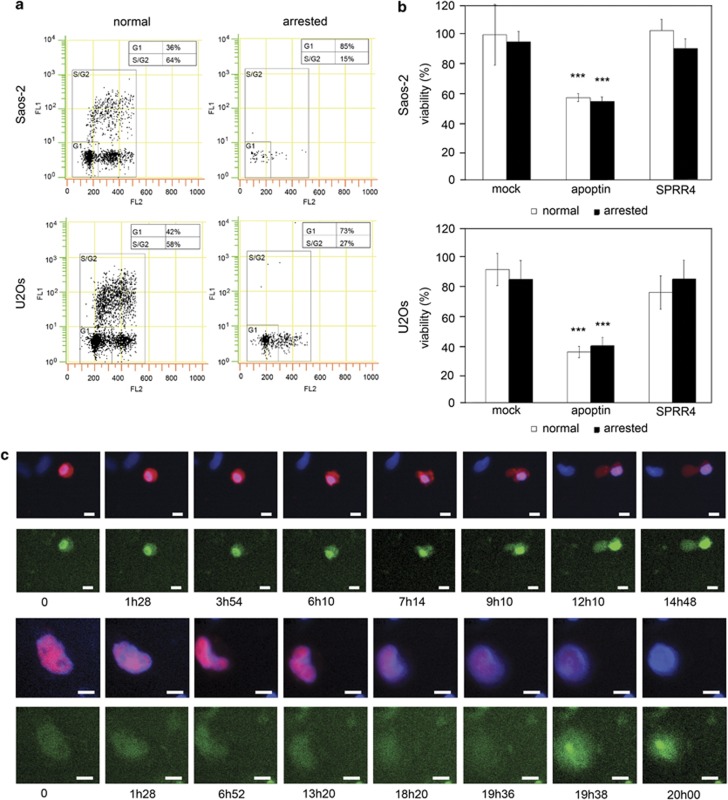
Figure 4.
Apoptin induces apoptosis in cycling and arrested Saos-2 and U2OS osteosarcoma cells. Cells were either grown under cycling conditions in normal culture medium, or arrested in G1/G0 by serum starvation and mimosine treatment. Bivariate dot-plots of a FACS analysis show the log distribution of the FITC anti-BrdU staining (FL1, green fluorescence, DNA synthesis) versus the linear distribution of PI staining (FL2, red fluorescence, DNA content). Percentages of gated cells in G1/G0 and S/G2 are indicated (a). Cells were transiently mock transfected or transfected with plasmid encoding either flag-apoptin or flag-SPRR4. Viability of both normal (white bars) and arrested cells (black bars) was measured by MTT assay (Saos-2 n=12, U2OS n=8). Under both situations, apoptin expression resulted in a significant decrease in cell viability (***=P<0.001) (b). Cell cycle-arrested U2OS cells transfected with plasmid encoding mCherry–apoptin (red) were incubated with NucView488 caspase-3 substrate (which turns green and binds to DNA when cleaved by activated caspase-3) and analyzed by time-lapse microscopy. Cells were incubated with Hoechst 33342 DNA stain (blue) prior to picture capturing. Time-lapse images were recorded for 20 h every 2 min 40–60 h post transfection. Filmstrips from two separate movies are shown. Time points of recording are indicated below the respective images. Overall, 29 out of 37 monitored apoptin-positive cells also stained for caspase-3 activity (78%). Scale bar: 10 μm (c)