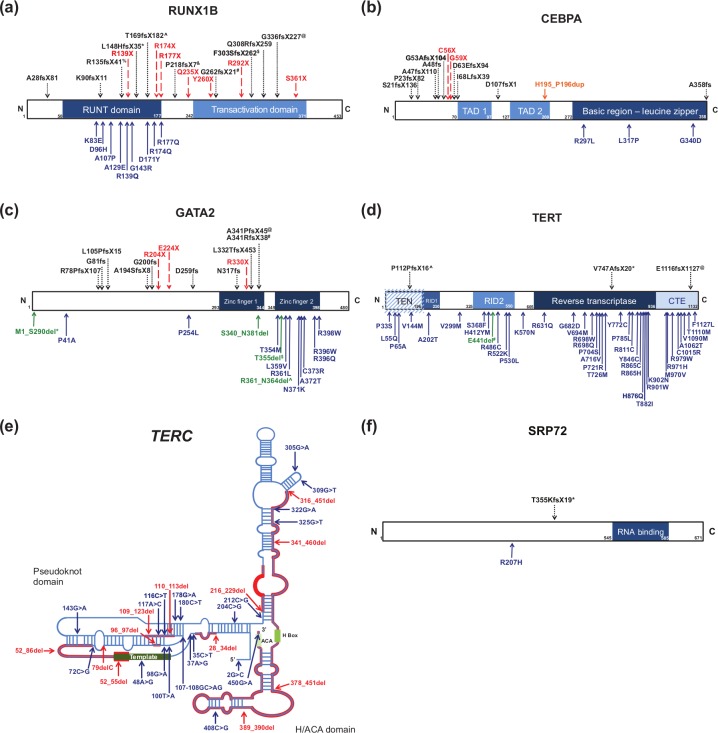
Figure 1.
Missense, nonsense, frameshift, duplication, and deletion mutations that confer familial predisposition to inherited leukemias. Missense mutations are shown in blue; nonsense mutations in red; frameshift mutations in black; duplications in orange; and deletions in green. Superscripts used for the deletion mutations delineate those shown in greater detail in Figure 2. (a) Protein schematic of RUNX1 isoform B (NP_001001890.1) (adapted from Owen et al. [2008b]). (b) Protein schematic of CEBPA (NP_004355.2) (adapted from Ho et al. [2009]). (c) Protein schematic of GATA2 (NP_116027.2) (adapted from Greif et al. [2012]). (d) Protein schematic of TERT (NP_937983.2) (adapted from Wyatt et al. [2009]). (e) Predicted RNA secondary structure of TERC RNA (NR_001566.1) (adapted from Vulliamy and Dokal [2008]).(f) Protein schematic of SRP72 protein (NP_008878.3) (adapted from Iakhiaeva et al. [2010]). CTE, C-terminal extension; RID, RNA interaction domain; TAD, transactivation domain; TEN, TERT essential N-terminal domain.