Zygomycetes surround us in our daily life, not only as agents of disease, but also as starters of fermentation in the preparation of food products, and as pioneer degraders in food spoilage. Since many members grow easily in axenic culture and show an impressive morphology, they have been subject of studies since the mid nineteenth century. Recent progress in whole genome sequencing projects aim to determine genetic features in genomic terms, such as their remarkable ability to pioneer on virgin substrates before competing microorganisms arrive. Although they exhibit a wide variety of different lifestyles including hyperparasitism and endosaprotrophy, many aspects of their ecology are still poorly understood. This diverse ecology may explain their changing faces: assisting humanity already over thousands of years in preparing our food, but also their relentless aggression towards the weakened human host.
During recent years zygomycetes have emerged as agents of disease in hospitalized patients. Presumably this is partly due to prophylaxis against Aspergillus fumigatus, which reduces the frequency of Aspergillus infections, and sees Mucorales fill a window of opportunity. Infections by species of Mucorales such as Lichtheimia, Mucor and Rhizopus may be highly mutilating in patients with severe leukemia, transplants, with ketoacidotic diabetes, or as agents of wound infections. Extended necrosis of tissue frequently leads to death in the susceptible patient population. The emergence of mucoralean fungi as agents of disease has resulted in a renewed global interest in zygomycetes. The present issue of Persoonia reflects the growing significance of this group of fungi.
Zygomycete research in the scientific community is presently stimulated and coordinated on an international basis by the ECMM-ISHAM (European Confederation of Medical Mycology, and International Society for Human and Animal Mycology) Working Group Zygomycoses, co-ordinated by George Petrikkos. The Working Group was founded in 2004 under ECMM. After organizing the 1st International Forum on Zygomycosis in 2008 at Cape Sounion, Greece, a supplement of Clinical Microbiology and Infection was published. A second Forum was held in 2010 at Porto Heli, Greece. A website was made in the same year (www.zygomyco.net) with a database where cases of mucormycosis can be submitted online. An overview of zygomycosis in Europe was published with 230 cases accrued by the registry of the Working Group (Skiada et al. 2011). The Working Group joined ISHAM in 2009. Several members of the group took part in a meeting in Chicago, organized by Thomas Walsh with the support of the Hank Schueler foundation and a special issue of Clinical Infectious Diseases was published (Walsh & Kontoyannis 2012). Subsequently a Special Interest Group (SIG) meeting was organized by Kerstin Voigt and Sybren de Hoog in conjunction with the International Mycological Congress on the Biology of Fungi (IMC9) held in Edinburgh, Scotland, August 2010. A follow-up meeting concentrating on zygomycete biodiversity took place in Utrecht, the Netherlands, March 2011. This meeting enabled fruitful discussions, inspiring an exchange of ideas, and providing an updated view on this group of fungi. The outcome of this meeting formed the basis of the present special issue of Persoonia.
Zygomycetes are among the most ancient fungi, and we aim to present a general overview of their impressive diversity. Starting with Krings et al. we provide insight into the fossil records of this fascinating group of fungi. We take a closer look at the largest order, the Mucorales, of which Walther et al. present a comprehensive molecular overview, Lu et al. describe a very remarkable representative of the order and Hoffmann et al. give an overview of family structures. Wagner et al. direct attention to the phylogeny of Mortierellales and Grygansky et al. present a phylogenetic overview of the Entomophthorales. Dealing with the problems of resolving the backbone in the fungal tree (see below), Tretter et al. evaluate the usefulness of some new loci for the resolution of basic lineages, particularly in the Kickxellomycetales.
Zygomycetes as agents of infection
Undoubtedly, the greater majority of fungi in the zygomycete amalgamate do not possess any virulence to mammals whatsoever. In clinical practice, the term ‘zygomycosis’ actually stands for infections by members of only three out of the 13 orders known, viz. the Mucorales, the Entomophthorales and the Mortierellales. Interestingly, pathologies observed in the three orders have hardly anything in common. Especially the syndromes of Mucorales and Entomophthorales in many ways are each other’s opposite. Infections by Lichtheimia or Rhizopus in the Mucorales typically are acute, which seems to be an expression of their pioneer ecology mentioned above. Angiotropism leads to rapid clotting of blood veins and extended tissue necrosis – with the enigmatic Mucor irregularis as the only exception (Li & Lun 2012). Infections by Basidiobolus or Conidiobolus in the Entomophthorales, however, are typically chronic, with hyphal elements residing in affected tissue over lengthy periods of time, leading to deposition of antigenic material around the fungus. The syndromes of Mucorales and Entomophthorales in many ways are each other’s opposite.
Do ‘zygomycetes’ exist at all, and can patients suffer from ‘zygomycoses’? In an influential paper, Hibbett et al. (2007) noted that the backbone of the phylogenetic tree of fungi traditionally united in the phylum ‘Zygomycota’ was not resolved, and thus that they do not compose a natural entity. The polyphyletic zygomycota should be abandoned, and hence the term ‘zygomycosis’ can no longer be applied for an infection caused by a member of that group. Disease names referring to the ordinal level would therefore be more informative than ‘zygomycosis’. ‘Entomophthoromycosis’ is available for infections caused by members of the Entomophthorales, while for those by Mucorales the term ‘mucoralomycosis’ would be logical. Although formally incorrect, the use of the more commonly used term ‘mucormycosis’ is advocated (Kwon-Chung 2012).
Taxonomic dilemmas at species level and above
With the recent technical improvements, molecular phylogeny is rolling over old concepts (Schoch et al. 2012). With taxonomies becoming more detailed, we observe a significant degree of rank inflation in the former zygomycetes. Many of the groups that classically were treated as orders, such as the Kickxellales or Zoopagales, today are raised to the status of the separate subphyla Kickxellomycotina and Zoopagomycotina. Some of these higher-level taxa contain only a very few species. Phylogenetically they are among the basal lineages of the fungal kingdom, a position ascribed to their early evolution in the history of life on earth (Berbee & Taylor 1993). By calibration of phylogenetic trees using fossil records with known geological age the ancestry of the zygomycetous groups was determined to lie at least a billion years back. Orders that are considered to be the most recent are roughly dated at 250 million years (Hoffmann et al. 2013). This implies that during times when higher ascomycetes started to emerge, the zygomycetous fungi were already fully adapted, and had successfully occupied their habitats. In many ways they seem to have remained unchanged. Hence zygomycete genera are mostly less speciose than many, more recently evolved ascomycetes, such as the dermatophytes where new taxa radiated during the last millenia (Gräser et al. 2006). It may be hypothesized that in ancient zygomycete species that survived over time the transcriptome has remained largely unaltered over millions of years, because many of the niches – such as competition-free, virgin materials – have universally been available over time. We speculate that their longer period of mutation and evolution is therefore reflected more in anonymous markers than in coding genes.
In contrast to morphology, datasets in phylogeny necessarily have a tree-like, fractal structure, the branch lengths being determined by phylogenetic age, mutation rates, as well as by processes of selection and adaptation. Translating the tree into a hierarchical taxonomic system, as was done rigorously by Hibbett et al. (2007) is not a priori better than the flat morphological system of the past. Degrees of change between taxa in introns, spacers and third codons, which are most widely applied in phylogenetic studies, are consistently larger in zygomycetous fungi than observed with comparable markers in most of the higher fungi. The overall higher degrees of molecular diversity is also reflected at the species level. For example, Alastruey-Izquierdo et al. (2010) observed 13 % ITS variability within an entity in Lichtheimia that with all parameters available at that moment should be regarded as a single species. Also in ancestral ascomycetous yeasts high degrees of intraspecific heterogeneity are known (Simon & Weiß 2008, Alper et al. 2011). In contrast, there are numerous complaints among workers in the top of the fungal tree that ITS may be invariable between species (Seifert et al. 2007, Chandra et al. 2011). Wouldn’t it be logical to allow different degrees of heterogeneity in different branches of the fungal tree of life? This might reinforce the role of biology, ecology and other basic traits as classification criteria compared to molecular phylogeny towards a more integrative taxonomy (Dayrat 2005, Padial et al. 2010). Time is the prime marker for phylogeny and evolution, but not necessarily for taxonomy. Besides, the prospect for novel universally applicable secondary barcodes is still not yet exhausted (Robert et al. 2011).
There is another, practical reason to refrain from drastic taxonomic decisions based solely on phylogenetic trees. Methods of tree reconstruction are becoming increasingly sophisticated, but in many sections of the fungal kingdom only a minor fraction of the extant diversity has been described. For example, Ruibal et al. (2008) grew fungi from crushed rock samples in Spain and found > 200 novel species; hardly any of the fungi they found was known. As a result of such significant additions, phylogenetic trees are subject to change, and this situation will certainly remain for the next decades at least, when poorly sampled habitats such as the intestinal tracts of amphibians, or drowned mosquitos in Siberian lakes have exhaustively been sampled.
Species delimitations in zygomycetous fungi on the basis of the biological species concept may be problematic when ascertained by mating experiments. Successful sexual interaction is a prime criterion to verify whether the high degrees of intraspecific variation observed in some of the fungi concerned truly match with species delimitations. However, sexual vigour may differ between strains, some being highly competent, showing response outside species limits. Alastruey-Izquierdo et al. (2010) observed zygospores between two suspensors, which just were smaller in shape and lighter in colour than those resulting from intraspecific mating. Geneological coalescence in silico may therefore be a more reproducible criterion to establish levels of intraspecific variation. Albeit difficult to define, the species is the only significant taxonomic category in fungi, because basically it exists in the real world and has defining criteria in genealogical concordance analysis. All other taxonomic entities are human-made.
Pandora’s box of species
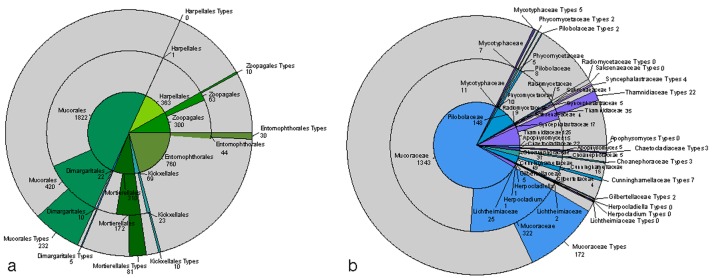
Despite all these hesitations, the zygomycetes presented in this issue of Persoonia may serve as a model for the growth of knowledge of diversity. Over one and a half century several paradigm shifts took place, such as culturing, elucidation of life cycles, and molecular phylogeny, which all left their footprints in the system of lower fungi. In zygomycetes, the use of living strains and deposition of type materials has been practiced as from the early days of mycology, in the mid-eighties of the 19th century. The CBS reference collection harbours a stunning number of names that are still represented by living ex-types (Fig. 1a), which can be investigated today with state-of-the-art technology. The number of reference strains held at CBS is very large compared to the number of species described in literature, particularly in the Mucorales (Fig. 1b). In the fungal kingdom this is only matched by the yeasts, which from their early days onwards were studied in culture using experimental methods. The availability of type materials of zygomycete species is essential now that numerous molecular siblings are being recognised, for which appropriate, validly described taxa may be hiding in the collection. Only since the publication of the International Code of Botanical Nomenclature in 1956, dried (metabolically inactive, i.e. in this case: dead) herbarium material was necessary for a valid description of a species; thus all names published before that time may be used. Authentic materials of these species should be re-evaluated by molecular means for eventual use of the name in state-of-the-art taxonomic systems. This will prevent the redescription of redundant, already existing taxa. Our study aims to stabilise nomenclature by including all available type material of zygomycetous fungi. The phylogenies with large datasets, such as those of Walther et al. (2013) and Wagner et al. (2013) show numerous taxonomic rearrangements compared to classical taxonomies of the same groups, and in this sense are revolutionary. Nomenclatural conservation is necessary for the widely used industrial, medical and model species. For many others name changes may be proposed, which do not severely impair the applied sciences. We hope that changes proposed after the appearance of this issue of Persoonia will be in line with our data and will become readily accepted.
Fig. 1.
a. Overview of the numbers of living strains of Zygomycetes held at CBS in each order. Inner circle: number of names in MycoBank, per order. Median circle: number of strains held relative to the number of names. Outer circle: number ex-type strains relative to the number of names. — b. Overview of the numbers of living strains of Mucorales held at CBS in each family. Inner circle: number of names in MycoBank, per family. Median circle: number of strains held relative to the number names. Outer circle: number of ex-type strains relative to the number of names.
One problem remains with reference materials for a number of species. For every new species, the Code requires that the author of the species deposits a dried or metabolically inactive specimen. If this was not done, and only the live strain was maintained, the living type is formally non-existent and the species is without a point of reference. As a result, without fulfilling these requirements the numerous authentic strains of zygomycetous fungi (Fig. 1) would not be usable as types for taxonomic studies. In this Persoonia issue, Walther et al. (2013) solved this dilemma by indicating the original, metabolically inactive, lyophilized material produced at deposition of the authentic strain as lectotype. If the strain was lyophilized many years after deposition – which was the case with a.o. all species described before the Second World War – an original drawing was selected as lectotype, and the living strain was designated as epitype. In that case, formally the nomenclatural status of the (for daily practice highly relevant) epitype is lower than that of the (practically useless) lectotype, but this solution gives us the possibility to treat living strains as types in phylogenetic trees, and thus nomenclatural stability is not jeopardized.