FIG 1 .
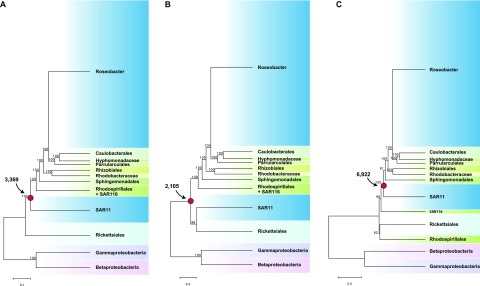
Model-based phylogenomic trees of alphaproteobacteria based on a concatenation of 60 orthologous protein sequences using the P4 Bayesian software with the NDCH and NDRH models (A), the RAxML software (B), and the PhyloBayes software with the CAT model (C). A Bayesian phylogeny using MrBayes with or without the covarion model had the same branching order as the RAxML tree. The node representing the most recent common ancestor (MRCA) of the Roseobacter and SAR11 lineages is indicated with a red dot, and the predicted gene number for the MRCA is indicated. For clarity, only the deep branches connecting the major lineages and their statistical support values are shown. The complete trees are shown in Fig. S1 in the supplemental material.
