FIG 1 .
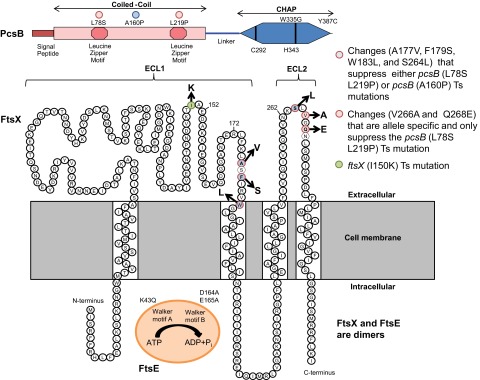
Summary of PcsB, FtsX, and FtsE domains and amino acid changes described in this paper. (Top) PcsB. Locations of changes in PcsBL78S-L219P(Ts) (red dots) and PcsBA160P(Ts) (blue dot) in CCPcsB and PcsBW335G(Ts) and PcsBY387C(Ts) in CHAPPcsB are indicated. Other structural features include the signal peptide, which is processed from the mature protein, the leucine zipper motifs in CCPcsB, and the required active-site amino acids Cys292 and His343 in CHAPPcsB (24). (Middle) FtsX. The predicted topology of FtsX is based on reference 19. Amino acid changes that suppress pcsB(Ts) mutations are color coded as indicated. The FtsXI150K change is also indicated. (Bottom) FtsE. The K43Q mutation and the D164A and E165A mutations in the Walker A and B motifs, respectively, are indicated. See the text for additional details.
