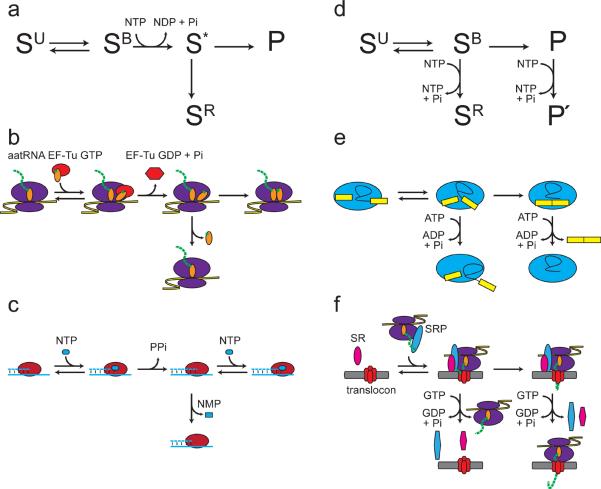
Figure 3.
Distinct manifestations of kinetic proofreading across diverse biochemical pathways. (a) Obligatory kinetic proofreading scheme employed during translation and DNA replication [8, 9]. Unbound substrate (SU) can engage the enzyme. The initial enzyme-substrate complex (SB) can dissociate or undergo an irreversible transition to an activated intermediate complex (S*) through the input of energy in the form of NTP hydrolysis. From the activated complex, conversion to a new on-pathway intermediate (P) competes with a branch in the pathway that results in rejection of the substrate (SR). (b) Kinetic proofreading during aminoacyl-tRNA selection on the ribosome [3]. Aminoacid-charged tRNA (green, amino acid; orange, tRNA) binds the ribosome (purple) as part of a ternary complex with EF-Tu and GTP (red). The initial recognition complex then undergoes irreversible GTP hydrolysis by EF-Tu, which then dissociates. In the subsequent inspection, accommodation of the aminoacyl-tRNA into the catalytic core of the ribosome where peptide bond formation occurs (preferred for cognate tRNAs) competes with dissociation of the activated aminoacyl tRNA (preferred for near-cognate tRNAs). (c) Kinetic proofreading during DNA polymerization [1]. An incoming dNTP (blue oval) forms base-paring interactions with the tempate strand (solid blue line) in the active site of DNA polymerase (red oval). The incoming dNTP is incorporated into the nascent strand (dashed blue line) through irreversible phosphodiester bond formation resulting in release of pyrophosphate. Base-pairing at the nascent 3' end of DNA is then inspected a second time as additional productive rounds of dNTP incorporation (preferred for a matched dNMP) compete with cleavage of the nascent phosphodiester bond after relocalization to the polymerase editing site (preferred for a mismatched dNMP), resulting in dissociation of dNMP (blue rectangle). (d) A distinct, non-obligatory kinetic proofreading scheme employed during splicing and protein translocation [8, 9, 16, 17, 39]. Unbound substrate (SU) binds the enzyme, forming an enzyme substrate complex (SB). From the enzyme–substrate complex, conversion to a new on-pathway intermediate (P) competes with an energy-dependent branch that results in substrate rejection (SR). However, if NTP hydrolysis occurs after the productive step then hydrolysis results in a new productive conformation (P'). (e) Kinetic proofreading during exon ligation [14]. At exon ligation, the lariat intermediate 3' splice site binds the spliceosome (blue oval) in a catalytic conformation. In this catalytic conformation, ligation of the exons (yellow rectangles) (preferred for optimal splice sites) competes with ATP hydrolysis by Prp22, resulting in rejection of the 3' splice site (preferred for suboptimal splice sites). ATP hydrolysis by Prp22 after exon ligation results in productive mRNA release [29, 30]. (f) Kinetic proofreading during protein targeting to the translocation machinery [10]. During protein targeting, a nascent signal peptide-ribosome complex is recognized by GTP-bound signal recognition particle (SRP, blue oval), which forms a complex with GTP-bound SRP receptor (SR, pink oval). At the translocon (red ovals), productive loading of the nascent peptide cargo into the translocation machinery (preferred for correct cargoes) competes with GTP hydrolysis by the SRP-SR complex, which results in dissociation of the cargo-SRP-SR complex (preferred for incorrect cargoes). GTP hydrolysis by SRP and SR after engagement of the cargo with translocation apparatus results in productive recycling of SRP and SR.