Figure I (Box 3).
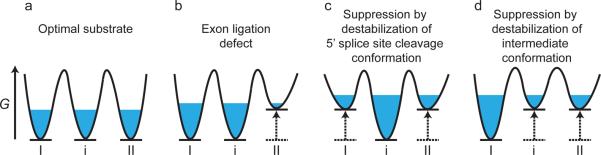
A three-state equilibrium model rationalizes genetic suppressors of substrate mutations. (a) In the model, a population of spliceosomes (blue) assembled on an optimal substrate equilibrates between 5' splice site cleavage (I) and exon ligation (II) conformations as well as an intermediate conformation (i). Note: free energies of each state are arbitrarily set to the same level. (b) The model implies that a mutation that impairs exon ligation destabilizes the exon ligation conformation relative to the 5' splice site cleavage and intermediate conformations, resulting in sequestration of the spliceosomes in these alternative conformations. (c and d) In the model, the exon ligation defect shown in (b) is suppressed by second-site mutations that repopulate the exon ligation conformation by destabilizing either the 5' splice site cleavage conformation (c) or the intermediate conformation (d), relative to the exon ligation conformation. Note that in all panels, destabilization of a particular state is illustrated as underlying changes in equilibria, but stabilization of alternative states would result in similar changes in equilibria. G, Gibbs free energy; in each panel the reaction coordinate proceeds from left to right.
