ABSTRACT
Quorum sensing (QS) regulates diverse and coordinated behaviors in bacteria, including the production of virulence factors, biofilm formation, sporulation, and competence development. It is now established that some streptococci utilize Rgg-type proteins in concert with short hydrophobic peptides (SHPs) to mediate QS, and sequence analysis reveals that several streptococcal species contain highly homologous Rgg/SHP pairs. In group A streptococcus (GAS), two SHPs (SHP2 and SHP3 [SHP2/3]) were previously identified to be important in GAS biofilm formation. SHP2/3 are detected by two antagonistic regulators, Rgg2 and Rgg3, which control expression of the shp genes. In group B streptococcus (GBS), RovS is a known virulence gene regulator and ortholog of Rgg2, whereas no apparent Rgg3 homolog exists. Adjacent to rovS is a gene (shp1520) encoding a peptide nearly identical to SHP2. Using isogenic mutant strains and transcriptional reporters, we confirmed that RovS/SHP1520 comprise a QS circuit in GBS. More important, we performed experiments demonstrating that production and secretion of SHP1520 by GBS can modulate Rgg2/3-regulated gene expression in GAS in trans; likewise, SHP2/3 production by GAS can stimulate RovS-mediated gene regulation in GBS. An isolate of Streptococcus dysgalactiae subsp. equisimilis also produced a secreted factor capable of simulating the QS circuits of both GAS and GBS, and sequencing confirms the presence of an orthologous Rgg2/SHP2 pair in this species as well. To our knowledge, this is the first documented case of bidirectional signaling between streptococcal species in coculture and suggests a role for orthologous Rgg/SHP systems in interspecies communication between important human pathogens.
IMPORTANCE
Pathogenic streptococci, such as group A (GAS) and group B (GBS) streptococcus, are able to persist in the human body without causing disease but become pathogenic under certain conditions that are not fully characterized. Environmental cues and interspecies signaling between members of the human flora likely play an important role in the transition to a disease state. Since quorum-sensing (QS) peptides have been consistently shown to regulate virulence factor production in pathogenic species, the ability of bacteria to signal via these peptides may prove to be an important link between the carrier and pathogenic states. Here we provide evidence of a bidirectional QS system between GAS, GBS, and Streptococcus dysgalactiae subsp. equisimilis, demonstrating the possibility of evolved communication systems between human pathogens.
Introduction
Bacteria coordinate gene expression among members of a community using sophisticated intercellular chemical signaling pathways in a process commonly referred to as quorum sensing (QS) (1). Since QS is a common mechanism used by pathogens to regulate virulence factor production and virulence-related behaviors, inhibitors of QS are now being considered as possible treatments for infections, especially in this time of increasing antibiotic resistance and treatment failures (2, 3). QS systems are currently best understood when bacteria are grown in pure culture; however, single bacterial species rarely exist in isolation either in the environment or within a host. It is therefore likely that such pure-culture studies have prevented the detection of interspecies signaling that, under natural conditions, may factor in to bacterial survival and/or pathogenesis.
Members of the genus Streptococcus are among the most prevalent types of bacteria found at sites throughout the human host (4). Many streptococcal species are known to be members of the normal, healthy human flora (5–8), while others are considered to be human pathogens but are also carried frequently in the host asymptomatically. In the vaginal tract in particular, many species of Streptococcus that can exist in both asymptomatic carriage and pathogenic states have been identified. A common member of the human vaginal flora is Streptococcus agalactiae (group B streptococcus [GBS]), with reported colonization rates between 6 and 36% (9–12). Vaginal carriage in women is typically asymptomatic but becomes particularly dangerous when the bacterium is transmitted from a pregnant woman to the fetus or neonate and is a leading cause of morbidity and mortality in newborns in the United States (13). Although best known as a neonatal pathogen and vaginal colonizer, GBS also colonizes many other human sites, including the skin, throat, and anorectum (14).
Another species capable of asymptomatic colonization at multiple sites within the host is Streptococcus pyogenes (group A streptococcus [GAS]). GAS is a major human pathogen responsible for a wide range of illnesses and life-threatening infections, such as strep throat, impetigo, toxic shock syndrome, and necrotizing fasciitis, as well as poststreptococcal sequelae like rheumatic fever and glomerulonephritis (15). Although GAS is predominantly regarded as a pathogen, numerous studies have demonstrated that 2 to 14% of school-age children asymptomatically carry GAS in the pharynx (16–20). Adult pharyngeal carriage rates are thought to be lower; however, reporting has been limited, and rates vary from ~2 to 10% (21, 22). Carriage of GAS has also been reported in the human vaginal tract, although at relatively low rates (23).
Until recently, very little was understood as to how or if pyogenic species of Streptococcus (e.g., GAS, GBS, Streptococcus dysgalactiae subsp. equisimilis, Streptococcus uberis, etc.) engaged in QS. It has recently been established that multiple streptococcal species, including at least one member of the pyogenic group, utilize proteins belonging to the Rgg family of transcriptional regulators to mediate QS activity facilitated by small-peptide pheromones (24–26). In 2011, we described a QS system conserved in all sequenced genomes of GAS (Fig. 1, left panel) (26). This system is mediated by two Rgg proteins, Rgg2 and Rgg3, which function as an activator and a repressor, respectively (Fig. 1). The rgg2 and rgg3 genes are each transcribed divergently from a gene encoding a short-hydrophobic-peptide (SHP) pheromone, shp2 and shp3, respectively. The shp genes, along with genes directly downstream of them, are regulatory targets of Rgg2 and Rgg3, resulting in a positive feedback loop characteristic of many QS pathways. Under noninducing conditions, the repressor Rgg3 binds the promoters of both shp genes, blocking transcription induced by Rgg2 via steric interference.
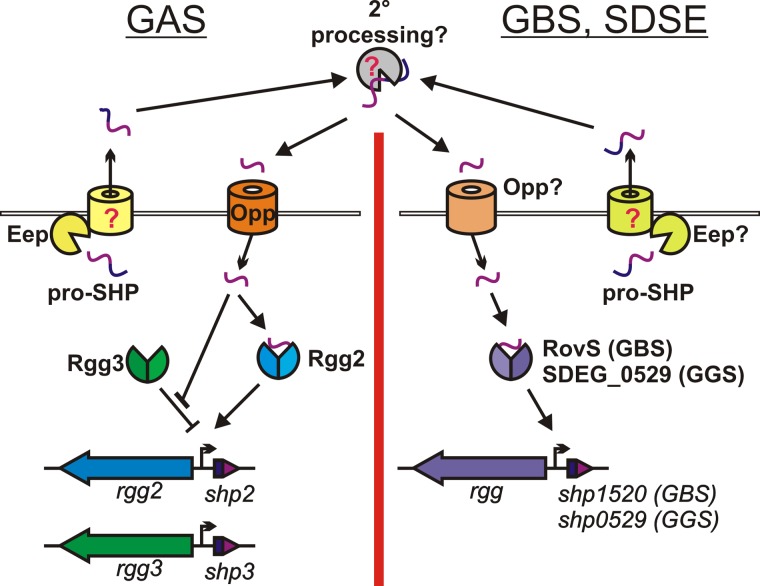
FIG 1 .
Proposed model for SHP-dependent interspecies signaling. In GAS (left), pro-SHP peptides, encoded by shp2 and shp3, are processed by Eep (26) and secreted by a yet-to-be-determined transporter. Secondary processing (proposed) produces a mature SHP pheromone that is imported by Opp (26). SHPs interact with Rgg3, disrupting transcriptional repression of shp promoters, and interact with Rgg2 to induce transcription of shp promoters. SHP production and response in group B and G streptococcus is proposed on the right half of the figure. Pro-SHP peptides encoded by rgg-adjacent genes are proposed to follow a secretory, processing, and importation pathway similar to that of GAS. RovS and SDEG_0529 are orthologs of Rgg2 and, at least in the case of RovS, display transcriptional activation properties described herein.
The metalloprotease Eep is required for cleavage of the pro-SHP peptides to their mature, active form comprised of the C-terminal 8 amino acids (SHP-C8), although the full mechanism of SHP maturation and export is currently unknown (26). When mature SHPs accumulate to sufficient concentrations in the extracellular environment, they are imported into the GAS cytoplasm in an Opp-dependent fashion (26), where they interact directly with both Rgg2 and Rgg3 (Fig. 1). Binding of SHPs to Rgg3 alleviates repression via disruption of DNA binding. The release of DNA from Rgg3 is thought to concede access of the shp promoters to activating Rgg2-SHP complexes, a requirement for robust transcription. In mutant strains lacking Rgg3, the system is constitutively on; in contrast, in mutant strains where Rgg2 has been genetically removed, the system remains off in the presence of SHPs even when Rgg3 is deleted. Thus, both the activator and repressor are required for accurate target gene regulation in GAS (26, 27).
Although GAS appears to be the only bacterial species that carries both the rgg2-shp2 and rgg3-shp3 pairs, several other species carry highly similar orthologs of either one pair or the other (Fig. 1, right panel). Orthologs of rgg2-shp2 are present in strains of GBS and S. dysgalactiae subsp. equisimilis, while rgg3-shp3 orthologs can be found in strains of Streptococcus porcinus, Streptococcus pneumoniae, and Streptococcus thermophilus (Table 1). Because of the high degree of similarity between the various Rgg orthologs, the high conservation of the C-terminal 8 amino acids comprising the putative active peptide, and the fact that multiple SHP2/3 carrying species are known to reside in the same location within the host, such as S. pneumoniae, S. dysgalactiae subsp. equisimilis, and GAS in the nasopharynx or GBS, S. dysgalactiae subsp. equisimilis, S. porcinus, and GAS in the female reproductive tract, we hypothesized that these Rgg/SHP pairs facilitate bidirectional communication between multiple species of streptococci.
TABLE 1 .
Rgg2/3 homologues and associated SHPs in select streptococcal species
| Species | rgg gene | Homology (%)a |
Binding siteb | SHP sequence | |
|---|---|---|---|---|---|
| Rgg2 | Rgg3 | ||||
| S. pyogenes NZ131 | spy49_0415 (rgg2) | 55/75 | 23/23 | MKKVNKAL-LFTLIMDILIIVGG | |
| S. pyogenes NZ131 | spy49_0449c (rgg3) | 55/75 | 23/23 | MKKISKFLPILILAMDIIIIVGG | |
| S. agalactiae A909 | SAK_1520 (rovS) | 79/87 | 53/74 | 20/23 | MKKINKAL-LFTLIMDILIIVGG |
| S. dysgalactiae subsp. equisimilis GGS_124 | SDEG_0529 | 85/92 | 53/74 | 23/23 | MKKINKAL-LLTLIMDILIIVGG |
| S. dysgalactiae subsp. equisimilis GGS-LT1 | Unannotated | 86/93 | 53/74 | 23/23 | MKKINKAL-LLTLIMDILIIVGG |
| S. thermophilus sp. CNRZ1066 | str1044 | 55/74 | 88/92 | 22/23 | MEKVSKILPILILVMDIIIIVGG |
| S. pneumoniae D39 | SPD_0939 | 55/74 | 90/95 | 22/23 | MKKISKFLPILFLVMDIIIIVGG |
| S. porcinus strain Jelinkova 176 | STRPO_0498 | 55/72 | 73/85 | 19/23 | MKEVLKSAPILLVIVDIIIIAGG |
Percent identity/similarity of gene product at the amino acid level to Rgg2 or Rgg3 of GAS.
Number of nucleotides conserved compared to the 23-nt Rgg binding site [TTTCCCACTTTC(A/C) AACAAAAA] identified in GAS (27).
To test this possibility, we examined two non-GAS species harboring Rgg2/SHP2 orthologs, namely, GBS strain A909 and S. dysgalactiae subsp. equisimilis strain GGS-LT1, for the ability to induce SHP-mediated QS in GAS. Both strains were indeed capable of inducing the Rgg2/3 system in GAS, and we proceeded to demonstrate that the Rgg2/SHP2 ortholog pair comprises a functional QS system in GBS that can respond to GAS-produced SHPs as well. The data presented herein provide proof of concept that several species of Streptococcus can both produce and respond to SHP pheromones originating with self, as well as those made by other species observed to carry an orthologous Rgg/SHP pair, thus enabling cross-species regulation of gene expression.
RESULTS
Comparison of Rgg2/3 homologs and associated shp genes in streptococcal species.
Recently, streptococcal genome analysis revealed a genetic linkage between genes encoding putative Rgg-type proteins and neighboring genes encoding putative small peptides (28). Further investigation has now demonstrated that these peptides function as cell-to-cell signaling pheromones via direct interaction with their cognate Rgg protein in multiple streptococcal species (24–26). Our lab has characterized one such system in GAS which utilizes two Rgg proteins, Rgg2 and Rgg3, in conjunction with two SHP pheromones, SHP2 and SHP3, to regulate biofilm biogenesis (26). Both of these Rgg/SHP pairs were previously assigned to group I of a phylogenetic tree based on Rgg protein similarity. This group contains Rgg proteins from several strains of other streptococcal species as well, including GBS, S. dysgalactiae subsp. equisimilis, and S. pneumoniae (28). Not included in the original analysis was S. porcinus strain Jelinkova, which also carries an Rgg/SHP pair with high homology to those of other group I members. The only group I Rgg protein other than Rgg2 and Rgg3 studied to date is SAK_1520 (named RovS), which was first described as a regulator of virulence genes in GBS (29). It was not until later that a systematic genomic report of rgg and shp genes identified a small open reading frame immediately adjacent to rovS encoding for a possible SHP peptide (28), and thus RovS has not yet been examined in the context of QS.
The protein sequence similarities between Rgg proteins of this group are all greater than 74% when compared to Rgg2 or Rgg3, with each protein sharing markedly more similarity with one Rgg protein than the other (Table 1). The SHP sequence similarity between strains also appears to correlate with the Rgg sequence similarity; Rgg proteins with greater similarity to Rgg3 have associated SHP pheromones more similar in sequence to SHP3 than to SHP2, and vice versa. This correlation can be most readily appreciated by comparing the C-terminal eight amino acids of the SHPs (SHP-C8), which vary by only a single amino acid between SHP2 and SHP3. The S. porcinus SHP associated with STRPO_0498 is an exception in that it carries the isoleucine characteristic of SHP3 but also contains an alanine in place of the valine which is conserved among the other members of this group. It should also be noted that another feature of the Rgg/SHP regions shared among all of the above strains is the 23-nucleotide (nt) conserved site identified as being required for DNA binding by Rgg2 and Rgg3 in GAS (27), suggesting that this site may function as the DNA recognition sequence for orthologous Rgg proteins as well. Interestingly, whereas all sequenced strains of GAS contain both Rgg2/SHP2 and Rgg3/SHP3, every other species containing a group I Rgg/SHP pair has only a single ortholog of one of these pairs, never both. For example, strains of GBS and S. dysgalactiae subsp. equisimilis carry orthologs of Rgg2/SHP2, whereas pneumococcal strains carry an Rgg3/SHP3 ortholog.
Separately, we note that Streptococcus macedonicus ACA-DC198 and Streptococcus infantarius subsp. infantarius CJ18, as well as Staphylococcus pseudointermedius E99, also carry orthologs of Rgg3/SHP3 (SMA_0156, Sinf_0147, and Spoe_1561, respectively); however, the predicted SHP3-like peptides of these strains are unique compared to SHP3 in ways that would be expected to decrease their effectiveness as a signal in other group I strains. For example, the mature SHPs predicted in S. macedonicus ACA-DC198 and S. infantarius subsp. infantarius CJ18 lack the terminal aspartic residue characteristic of the group I SHP-C8 peptides and known to be important for activity in GAS (26).
The GAS Rgg2/3 QS system can respond to secreted signals produced by other streptococcal species.
Given that several strains of other streptococcal species carry shp genes that encode C-terminal ends that are the same as or highly similar to those of SHP2 and SHP3, we hypothesized that such strains may be able to cross-activate the Rgg2/3 system of GAS via production of orthologous SHPs. Thus, we sought to examine if spent culture supernatants of species carrying Rgg/SHP orthologs could induce a Pshp2 luciferase reporter known to be responsive to SHP-C8 peptides in GAS (26). To this end, we generated a GAS strain deficient in SHP pheromone production by mutating the translational start codons of shp2 and shp3 to GGG (shp2GGG and shp3GGG). We then introduced the Pshp2 reporter into this strain, generating strain BNL177, which allowed us to examine the QS-inducing activity of donor spent supernatants without interference by endogenously produced pheromone. Given that in GAS, Rgg2 functions as an activator and Rgg3 functions as a repressor, we decided to focus on species that contain an Rgg2/SHP2 ortholog pair, namely, GBS strain A909 and S. dysgalactiae subsp. equisimilis strain GGS-LT1. Such strains would be predicted to be “on” given the presence of a putative activator and lack of repressor. GGS-LT1 has not been fully sequenced and annotated, but PCR analysis and sequencing of the orthologous rgg-shp region of this strain revealed 99% identity of the rgg and shp genes to those of the sequenced S. dysgalactiae subsp. equisimilis strain GGS_124, as well as conservation of the putative Rgg binding site (data not shown).
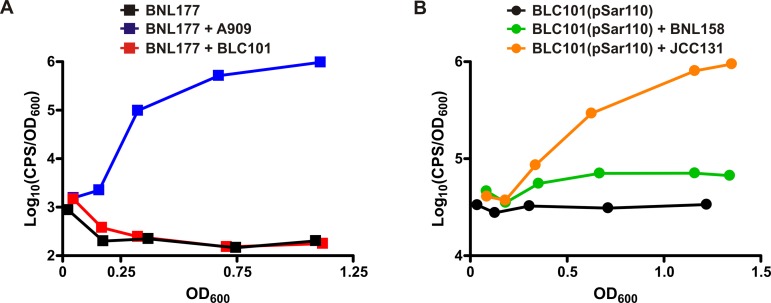
To examine if GBS strain A909 or S. dysgalactiae subsp. equisimilis strain GGS-LT1 produced a secreted factor capable of stimulating the Rgg2/3 system of GAS, cell-free spent culture supernatants were tested for their ability to induce the reporter strain BNL177. As negative and positive controls, respectively, supernatants from wild-type (WT) GAS and an isogenic Δrgg3 mutant were also included, since the QS system is known to be off in WT GAS but constitutively on in the Δrgg3 mutant (26). As expected, no reporter induction was detected when BNL177 was grown in fresh medium or in spent supernatant from WT GAS, whereas spent supernatant from the GAS Δrgg3 mutant induced reporter expression (Fig. 2A). More importantly, spent supernatants from both GBS and S. dysgalactiae subsp. equisimilis induced robust reporter expression to levels even greater than that of the positive control (Fig. 2A). These results were the first evidence that a secreted factor produced by GBS A909 and S. dysgalactiae subsp. equisimilis GGS-LT1 was indeed capable of inducing shp gene expression in GAS.
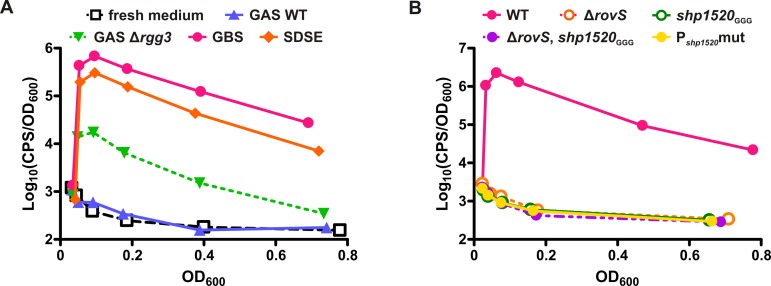
FIG 2 .
GAS responds to signals produced by other species, and the GBS signal is dependent on RovS and SHP1520. A shp-null GAS strain containing a Pshp2 –lux reporter, BNL177, was grown to log phase and diluted in spent culture supernatants from various strains. Luminescence expression indicates induction of the SHP system in GAS. (A) SHP2 expression is activated by spent culture supernatants from WT GBS (A909) and S. dysgalactiae subsp. equisimilis (GGS-LT1), as well as a Δrgg3 GAS strain (JCC131) that produces high levels of SHP peptide. Fresh medium and WT GAS (NZ131) supernatant do not induce the reporter. (B) Production of the secreted inducer in GBS supernatant is dependent on RovS, SHP1520, and the putative RovS binding site. Spent supernatants from GBS strains with mutations in rovS (BLC100), shp1520 (BLC101), both rovS and shp1520 (BLC102), or the putative RovS binding site (Pshp1520mut; BLC103) are unable to induce luminescence expression in BNL177, unlike the WT A909 parent strain. Data are shown as a function of relative light units (CPS/OD600) versus OD600. Each graph is representative of at least 3 biological replicates.
RovS and SHP1520 are required for the GAS-inducing activity of GBS.
We next sought to confirm the identity of the secreted factor(s) inducing the BNL177 reporter strain, which we hypothesized to be the SHP2 orthologs. Given that A909 has been fully sequenced and is amenable to genetic manipulation, we decided to focus on this strain for further investigation of interspecies signaling. To determine if the SHP2 ortholog of A909, which we have named SHP1520 due to its proximity to the RovS-encoding gene SAK_1520, was responsible for reporter induction in BNL177, we generated several isogenic A909 mutant strains in which we mutated genetic components predicted to be important for SHP1520 production. We hypothesized that not only would the shp1520 gene itself be required for peptide production, but RovS would likely also be important given that Rgg2 is required for robust shp2 expression in GAS. We generated an internal deletion in rovS (ΔrovS; BLC100), a mutation of the shp1520 translation start codon to GGG (shp1520GGG; BLC101), and a double mutant in which rovS and shp1520 were both mutated (ΔrovS shp1520GGG; BLC102). We then tested spent culture supernatants from these isogenic mutant strains of A909 for inducing activity, again using the GAS reporter strain BNL177. Whereas WT A909 supernatants induced reporter expression, none of the supernatants from mutant strains had any effect on Pshp2 reporter expression (Fig. 2B). These results confirmed that rovS and shp1520 are required for production of the secreted factor responsible for BNL177 reporter induction, supporting our hypothesis that SHP1520 itself is the activating signal and that RovS is involved in SHP1520 production.
The DNA binding site of Rgg2 and Rgg3 that is required for Rgg-mediated regulation of shp gene expression in GAS is conserved in A909. We therefore hypothesized that this regulatory element may also be essential for shp1520 expression in A909. If the 23-nt conserved site functions analogously in GBS, mutation of the putative binding site would be expected to abolish SHP1520 production and subsequent induction activity. Indeed, when we mutated two nucleotides within the conserved 23-nt site (Pshp1520mut; BLC103), there was no longer any inducing activity in the spent supernatants despite intact rovS and shp1520 genes (Fig. 2B). This not only confirms the requirement for SHP1520 in cross-species induction of the Rgg2/3 system but also supports our hypothesis that RovS is involved in activation of shp1520 expression and does so via interaction with the same regulatory element used by Rgg2 and Rgg3 in GAS.
RovS and SHP1520 comprise a QS circuit in GBS.
Many QS systems possess an autofeedback loop in which the signaling molecule positively regulates its own expression at the transcriptional level (1). In all Rgg/SHP QS systems studied to date, the Rgg protein regulates expression of the adjacent shp gene (24, 26, 30–32). To examine if RovS and SHP1520 comprise a functional QS system in GBS, we generated transcriptional reporters in which the putative promoter region of shp1520 alone or the shp1520 promoter region in combination with the entire rovS gene were fused to bacterial luxAB genes, generating the reporter plasmids pSar110 and pSar111, respectively (Fig. 3A). These reporters allowed us to directly assess shp1520 gene expression levels by measuring luciferase production from GBS reporter strain cultures. We transformed these reporters into WT A909 and into the isogenic mutants in which rovS, shp1520, or both genes were mutated and compared luciferase production to examine if RovS and/or SHP1520 contributed to the regulation of shp1520 gene expression.
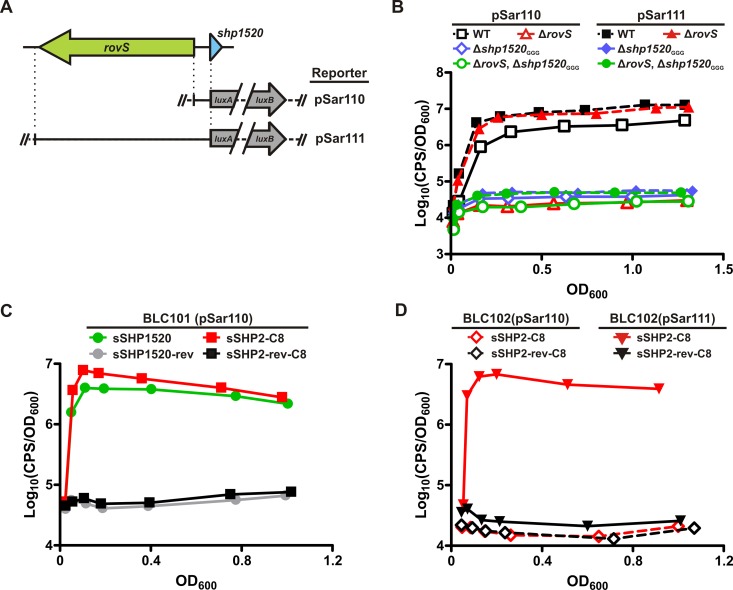
FIG 3 .
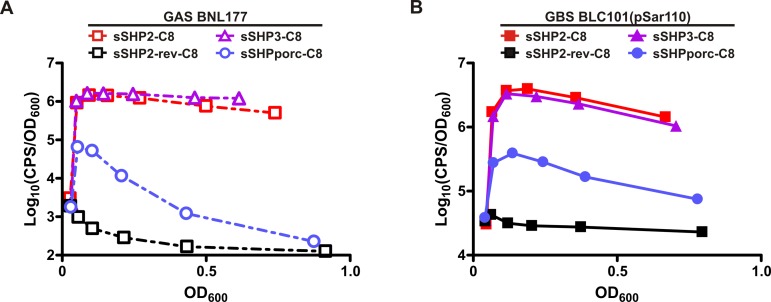
RovS and SHP1520 comprise a functional QS system in GBS. Regulation of Pshp1520 is mediated by both RovS and SHP1520. (A) Schematic diagram of DNA fragments used in transcriptional fusions with luxAB for use in GBS strains. Both reporters contain the putative shp1520 promoter, but pSar110 omits the rovS gene, whereas pSar111 includes rovS in its entirety. (B) Expression of Pshp1520 (pSar110) is autoinduced in cells containing both RovS and SHP1520 (A909). In strains lacking rovS (BLC100), shp1520 (BLC101), or both genes (BLC102), Pshp1520 is not induced. Additional copies of rovS on a plasmid (pSar111) complement the ΔrovS strain (BLC100) and slightly increase induction levels in the WT strain (A909). (C) Exogenous synthetic full-length SHP1520 peptide, as well as C8 peptide but not reverse peptides, induces Pshp1520 expression in BLC101(pSar110), thus complementing the shp1520 mutation. (D) sSHP2-C8 but not the reverse peptide induces Pshp1520 expression in a RovS-dependent manner. Data are shown as a function of relative light units (CPS/OD600) versus OD600. Each graph is representative of at least 3 biological replicates.
In WT A909, luminescence from pSar110 was induced as the cells entered logarithmic phase and remained high throughout growth (Fig. 3B). When rovS was included in the plasmid (pSar111), luminescence came on earlier and was sustained at a higher level, presumably because RovS was expressed to higher levels. In contrast to WT cultures, there was no detectable luminescence from pSar110 in any of the mutant strains, consistent with the lack of inducing activity in spent supernatants from these strains. Additionally, inclusion of rovS on the reporter plasmid could rescue luminescence induction in the ΔrovS mutant (BLC100) but had no effect in the shp1520GGG (BLC101) or double knockout ΔrovS shp1520GGG (BLC102) strains (Fig. 3B).
To ascertain if the absence of SHP1520 was responsible for the inability of native or overexpressed RovS to activate Pshp1520 reporter expression in BLC101, we examined luminescence when a synthetic SHP1520 peptide was added to the reporter cultures. We tested both full-length SHP1520 and a truncated version comprised of the C-terminal eight amino acids, since the C8 portion of SHP2 and SHP3 comprises the active signaling molecule in GAS. Because the C8 portions of SHP2 and SHP1520 are identical (DILIIVGG), we will refer to this synthetic C8 peptide as sSHP2-C8.
In agreement with a requirement for SHP1520 in RovS-mediated activation of SHP1520 expression, the sSHP1520 peptide rescued induction from the reporter in BLC101 with or without RovS overexpressed from the plasmid (Fig. 3C). This was expected given that this strain harbors an intact rovS gene in the chromosome. As a control, a full-length synthetic peptide with the reverse sequence (sSHP1520-rev) was also tested but had no effect on reporter induction. sSHP2-C8 was also able to induce expression from the GBS reporter, whereas a reverse peptide (sSHP2-rev-C8) was not (Fig. 3C), indicating that GAS and GBS can recognize the same mature form of the peptide.
To confirm that SHP1520 functions to induce its own expression solely via RovS, we tested the ability of sSHP2-C8 to induce reporter expression in BLC102 with or without rovS expressed from the reporter plasmid. In the absence of rovS (pSar110), there was no reporter induction detected in the presence of either sSHP2-C8 or the control reverse peptide (Fig. 3D). In contrast, when rovS was included in the reporter plasmid (pSar111), sSHP2-C8 was able to induce robust reporter expression, whereas the reverse peptide had no effect. Thus, in agreement with other Rgg/SHP QS systems, the SHP1520 pheromone (likely the C8 portion of the molecule based on successful induction with synthetic C8 peptide) functions to induce its own expression in a RovS-dependent manner, and RovS can activate expression from the shp1520 promoter only in conjunction with the SHP1520 pheromone.
The GBS QS system can respond to SHPs produced by other streptococcal species.
Based on the confirmed functions of RovS and SHP1520 in a QS circuit and the finding that GAS responds to SHP peptides produced by GBS and S. dysgalactiae subsp. equisimilis (Fig. 2A), we predicted that GBS would also respond to SHPs produced by other streptococcal species. To test this, we used the GBS shp1520GGG reporter strain carrying the Pshp1520 reporter plasmid [BLC101(pSar110)]. Since this strain cannot produce its own SHP pheromone but carries an intact copy of rovS, spent culture supernatants should be able to induce reporter expression if they contain a secreted SHP peptide recognizable by RovS. Consistent with the ability of SHP2-like peptides to cross-activate strains carrying an Rgg2 ortholog, spent supernatants from JCC131 (GAS Δrgg3 mutant in which shp expression is robust) and GGS-LT1 cultures were able to induce high-level reporter induction compared to results with fresh medium (Fig. 4). Surprisingly, spent supernatant from the negative-control strain NZ131 was able to slightly induce reporter expression compared to fresh medium (Fig. 4), albeit far below the levels induced by JCC131 or GGS-LT1. This slight induction may represent low-level expression of SHPs in WT GAS that was previously undetected (26) because of the single-copy nature of the GAS transcriptional reporters as opposed to the multicopy plasmid GBS reporter. Alternatively, the concentration of SHP that effectively activates RovS may be lower than that needed for Rgg2/3 activation. Further studies will be needed to investigate these possibilities.
FIG 4 .
GBS responds to other bacterially derived SHPs. An SHP-deficient strain of GBS (BLC101) carrying a Pshp1520 reporter (pSar110) is highly induced in response to spent culture supernatants from GAS Δrgg3 (JCC131) and S. dysgalactiae subsp. equisimilis (GGS-LT1) strains but not in response to fresh medium or WT GAS (NZ131) supernatant. Data are shown as a function of relative light units (CPS/OD600) versus OD600. The graph is representative of at least 3 biological replicates.
As previously mentioned, GAS is the only strain known to contain both Rgg2/SHP2 and Rgg3/SHP3 pairs. However, the active portions of the two SHPs are identical at 7 of 8 positions. We wondered if a species containing only an Rgg2/SHP2 ortholog pair would still be able to respond to SHP produced from a species containing only an Rgg3/SHP3 pair. Additionally, we wondered if the predicted C8 portion of the SHP pheromone from S. porcinus, in which the conserved valine is replaced by an alanine, would have activity in Rgg2-containing strains. To this end, we tested reporter induction in both GAS BNL177 and GBS BLC101 (pSar110) in response to synthetic versions of sSHP2-C8, sSHP3-C8, and S. porcinus C8 (sSHPporc-C8). The sSHP2-C8 and sSHP3-C8 peptides were able to induce high-level induction in both reporter strains (Fig. 5A and B). Each reporter strain also responded to the sSHPporc-C8 peptide, though the level of induction was lower than that seen with either of the other peptides. Although a fairly minor substitution, the valine-to-alanine change may result in slightly weaker interaction with the Rgg proteins, causing the observed lower level of induction. Overall, these data suggest that all strains belonging to group I on the Rgg protein phylogenetic tree can likely cross-signal by virtue of the highly similar C-terminal ends of their SHP peptides.
FIG 5 .
Both GAS and GBS can respond to nonnative synthetic SHP-C8 peptides predicted to be produced by other species. SHP-null GAS reporter strain BNL177 (A) and GBS reporter strain BLC101 (pSar110) (B) are both induced by sSHP2-C8, sSHP3-C8, and sSHPporc-C8 but not by sSHP2-rev-C8. sSHPporc-C8 induces both reporters approximately 10-fold less than sSHP2-C8 or sSHP3-C8. Data are shown as a function of relative light units (CPS/OD600) versus OD600. Each graph is representative of at least 3 biological replicates.
Native SHP production can cross-regulate gene expression during coculture.
Although the data presented above indicate that bacterially produced SHP peptides can function to induce gene expression in heterologous species, all experiments were done by cross-feeding with cell-free spent culture supernatants or synthetic peptides. Given that some streptococcal strains have been postulated to exert inhibitory growth effects on one another (33), we felt it was important to test whether SHP-mediated interspecies signaling can occur when multiple SHP-producing species are growing together. To this end, we undertook coculture experiments in which shp-deficient GAS or GBS reporters were grown together with donor strains that were either competent or deficient in SHP production. Reporter luminescence and growth of both species were monitored over time. Given that the species carrying the reporter is unable to produce its own SHP but can still respond to SHP via its intact Rgg regulator, luminescence should be induced only when SHP is provided in trans by an SHP-producing donor strain.
The SHP-deficient GAS reporter strain (BNL177) was mixed at an approximately 1:1 ratio with GBS strains that either constitutively produce (A909) or do not produce (BLC101) the SHP1520 peptide. As expected, the BNL177 reporter turned on in the presence of A909 but not in the presence of BLC101 (Fig. 6A). When the opposite experiment was performed, in which the SHP-deficient GBS reporter strain [BLC101 (pSar110)] was mixed with either JCC131 (Δrgg3; SHP is expressed at high levels) or BNL158, (WT derivative in which SHP expression is repressed), SHP-dependent induction of the reporter was observed. JCC131 was able to induce strong reporter expression in BLC101(pSar110), whereas BNL158 was not (Fig. 6B). In agreement with earlier results (Fig. 4), very low level induction was seen in the GBS reporter when grown with BNL158 (Fig. 6B) despite the fact that shp expression is known to be repressed in this strain (26). Importantly, CFU counts taken during coculture growth showed no inhibition of growth of either GAS or GBS (data not shown).
FIG 6 .
Native SHP production can modulate cross-species gene expression when grown in coculture. (A) GAS reporter strain BNL177 was mixed with GBS strains that either produced (A909) or did not produce (BLC101) SHP1520. Coculture with A909 but not BLC101 induced expression of the Pshp2 reporter. BNL177 cultured alone was included as a control. (B) The GBS reporter strain BLC101(pSar110) was mixed with GAS strains in which the Rgg2/3 system is known to be highly induced (JCC131) or not induced (BNL158). Coculture with JCC131 but not BNL158 induced robust expression of the Pshp1520 reporter. BLC101(pSar110) cultured alone was included as a control. Coculture did not detectably inhibit growth of either GAS or GBS compared to growth in pure culture (data not shown). Data are shown as a function of relative light units (CPS/OD600) versus OD600, with the OD600 values being representative of total culture density. Each graph is representative of at least 3 biological replicates.
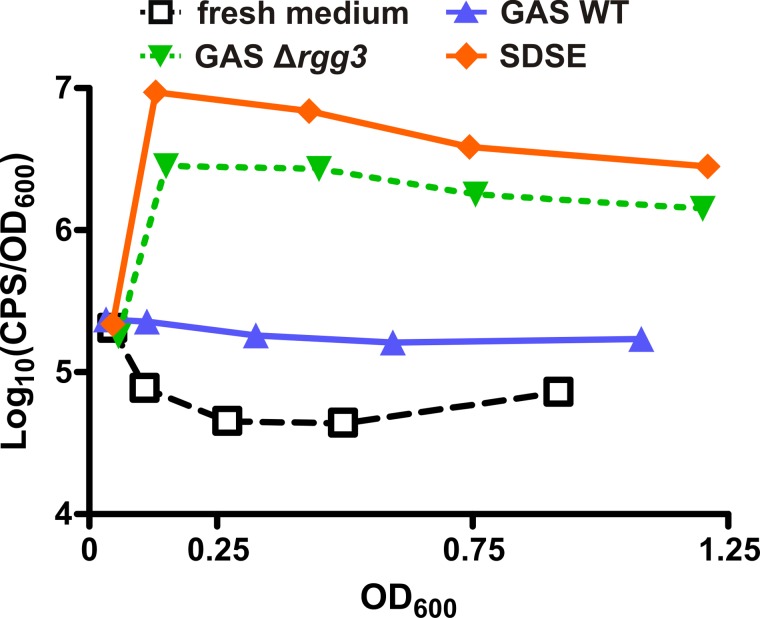
GBS-produced SHP increases biofilm formation in GAS.
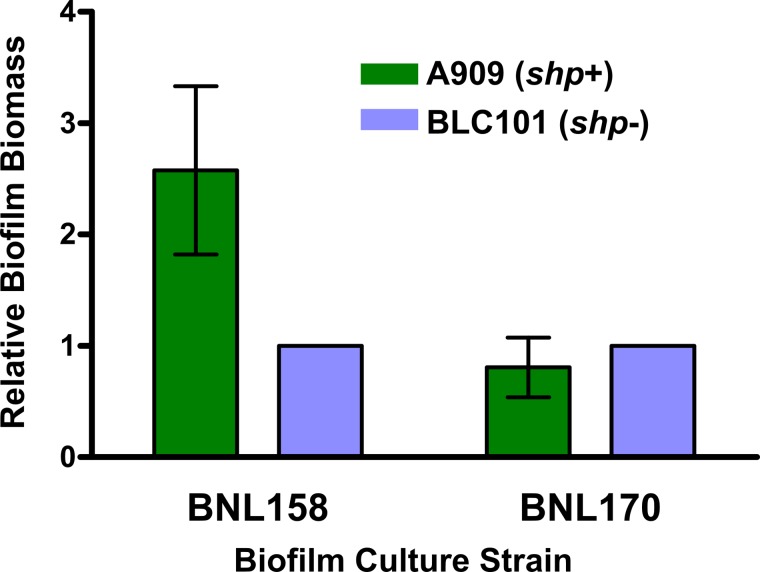
It has been previously demonstrated that induction of the Rgg2/3 circuit in GAS results in enhanced biofilm biogenesis (26). Thus, we sought to determine if heterologous SHP produced by GBS would be able to cross-modulate this phenotype in GAS. To this end, we measured biofilm growth of WT or shp-deficient GAS (BNL158 and BNL170, respectively) when grown in conditioned supernatants of GBS strains that produce (A909) or do not produce (BLC101) SHP1520. In agreement with our hypothesis, WT GAS (BNL158) biofilms grown in SHP-containing supernatants formed biofilms with increased biomass compared to those grown in supernatants from a shp-deficient donor strain (Fig. 7). A GAS strain incapable of producing SHP peptides did not show increased biofilm biomass when grown in SHP-containing medium (Fig. 7). This was not unexpected, since these cells cannot activate the positive feedback loop and thus expression of shp-induced genes rapidly decreases after initial activation. These results demonstrate that SHP-mediated cross-induction of gene expression can result in modulation of associated phenotypes in at least one direction and suggest a role for interspecies signaling in biofilm formation.
FIG 7 .
Heterologously produced SHP stimulates GAS biofilm production. Biofilm biomass of WT (BNL158) and shp-deficient (BNL170) GAS strains grown on aclar membranes was measured by enumeration of viable CFUs following 24-h incubation in the presence of spent culture supernatants from GBS strain A909 (WT) or BLC101 (shp1520GGG). Levels of biofilm production for each GAS strain were normalized to that of the same strain grown in spent supernatants from BLC101, and error bars indicate standard errors of the means (n = 3).
DISCUSSION
Metagenomic analyses of the human microbiome have made it possible to relate the composition of microorganisms residing in the human body to the health and disease of the host. Early studies support the idea that structure of the microbiome has a critical impact on health (6), and it is anticipated that these findings will assist in understanding how and why some bacteria transition from benign occupant to virulent pathogen. Considering the diversity and large numbers of bacteria present at many sites in the human host, it seems highly unlikely that bacteria disregard the existence of unrelated microbes with which they coexist, compete, or possibly cooperate. Very little understanding currently exists as to how different species of the microbiome influence one another. The demonstration presented herein that not only is interspecies communication feasible but a genetically conserved pathway is present among species comprising members of the human flora raises questions regarding how such interactions contribute to health and disease. An understanding of these questions may allow preservation of health by preventing potential pathogens from transitioning from an asymptomatic to a pathogenic state.
Streptococci are prominent members of the human microbiota at several sites in the body, including the throat, nares, mouth, vaginal tract, skin, and anorectum. Oral streptococci have offered the best model for studying intercellular signaling, and several cases of cross-species signaling have been documented. In one case, two mutualistic species of bacteria (Streptococcus oralis and Actinomyces naeslundii), which are among the founding species of dental plaque, were shown to rely on the small signaling molecule known as AI-2 or DPD (di-hydroxy pentanedione) in the process of developing biofilms (34). DPD is a by-product of the reactive methyl pathway (35) and is an interspecies signal that reflects the metabolic status of a bacterial cell (36). Composition, structure, and mass of the dual-species biofilm were dependent on a narrow concentration of DPD produced by members of the biofilm, in particular that produced by S. oralis (34). In a different study, a peptide-mediated QS system common in strains of Streptococcus mutans and known to control bacteriocin production was found to be inhibited by other mitis-group streptococci, possibly through the degradative action of secreted proteases (37). Though these reports provided the first demonstrations that streptococci participate in interspecies communication, these examples only reported unidirectional signaling in cocultures, where one species elicited or blocked responses in another, but did not present evidence that there was a capacity for dialog.
GAS and GBS are often found at the same sites within the human host, and genomic studies have shown that lateral gene transfer (LGT) likely occurs between them, providing further evidence these species coexist and possibly interact (38–41). Interaction among other streptococci, including S. dysgalactiae subsp. equisimilis and S. porcinus, both of which can be found in host sites in which GAS and GBS are known to reside (42–45), also seems likely. In this study, we provide evidence that SHP signaling pheromones can facilitate bidirectional interaction between different streptococcal species, including GAS, GBS, and S. dysgalactiae subsp. equisimilis. The effect of this signaling can be observed as stimulation of promoters expressing pheromone genes in heterologous species, setting up a feed-forward or positive feedback loop. We have demonstrated that RovS and SHP1520 comprise a QS circuit in GBS reminiscent of the Rgg2/3 system of GAS save for the lack of a repressor akin to Rgg3. Further experiments will be needed to identify the maturation, export, and import pathways involved in SHP-mediated QS in GBS, but we hypothesize that homologues of the processing enzyme Eep and the oligopeptide permease Opp responsible for SHP import in GAS (26) could function analogously in the RovS/SHP1520 circuit of GBS and/or other Rgg/SHP ortholog-containing species (Fig. 1).
Finally, our data also indicate that interspecies signaling can regulate biofilm formation in GAS (Fig. 7). Further research will indicate whether this phenomenon also affects GBS biofilm formation or other virulence-related phenotypes and whether these SHP signals could affect the switch from colonization to pathogenicity by controlling attachment and biofilm formation in vivo. Overall, these data support the idea that SHP-based communication provides a means to influence multispecies consortiums in vivo.
One obvious difference between the SHP/Rgg quorum-sensing systems in GAS and those in other group I Rgg-containing species, including GBS, is the composition of regulatory proteins and the observed state of SHP expression when grown in a chemically defined medium. In GAS, the presence of a negative regulator (Rgg3) keeps the system off unless SHP pheromone is supplied to the culture, either in the form of active spent culture supernatants or synthetic peptide. GBS, on the other hand, does not contain an Rgg3 homolog, and expression of shp1520 is observed to be uninhibited under these growth conditions. While it cannot be assumed that high expression occurs in the body, if SHP production by GBS is capable of reaching a concentration at which GAS is able to respond, this may indicate that GBS provides a means to trigger the SHP positive feedback loop within GAS, thus promoting a consensus between these species.
The definitive behavioral changes induced by the Rgg2/3 QS system are not fully understood in either GAS or GBS. In GAS, induction of the system has been shown to lead to increased biofilm formation (26), and in GBS, RovS has been implicated in the control of virulence factor production (29). In addition to shp genes being induced by these circuits, we have found that the genes downstream of each shp gene in GAS are highly induced as well. This includes approximately 10 kb of coding sequence of unknown function downstream of shp3 and 1.5 kb downstream of shp2 (26; unpublished results). Further studies in elucidating the genetic responses to SHPs will be a priority in determining how each organism benefits from this signaling pathway and for testing the potential roles that pheromones play in carriage, persistence, and virulence of these species and in other Rgg/SHP-carrying pathogens. Consideration should also be given to the possibility that interspecies signaling may serve as a means to deceive, divert, or dissuade competing bacteria from carrying out decisions relying on QS, and coculturing may be required to reveal such antagonistic relationships.
Members of the Rgg protein family are conserved across the order Lactobacillales, and studies continue to investigate and define their role in QS. Relationships among bacteria in complex microbial ecosystems, like that in the human body, may become easier to define if lines of communication between species are recognized, and Rgg/peptide orthologs constitute a logical starting point for such investigations. Many other Rgg variants are recognized among bacterial genomes, and in some cases, small SHP-like peptides are predicted (24). For instance, another Rgg/pheromone system, ComR/ComS, is required for genetic transformation in S. mutans (25) and S. thermophilus (31) and was found to regulate competence-related genes in GAS (30). ComR and ComS are conserved among all species of the pyogenic, bovis, mutans, and salivarius groups (but not in mitis or anginosus species), and it has been demonstrated that synthetic ComS pheromones of heterologous streptococci were capable of ComR-dependent activation in S. thermophilus (46). The genetic potential for microbial communication begins to offer an extraordinary image that peptide-based interspecies communication is a complex network of pheromone varieties originating with numerous members of the community.
Moving toward an understanding of communication in the human microbiome as a whole will begin with expanding our knowledge of multispecies interactions. Since the ability of organisms to communicate via QS has been implicated in many virulence-related behaviors, understanding how organisms communicate and how cross-species communication affects virulence and persistence of pathogenic bacteria will allow us to develop new potential treatments that interfere with QS pathways, conceivably lessening the morbidity and mortality associated with bacterial infections.
MATERIALS AND METHODS
Bacterial strains and media.
All strains used in this study are listed in Table 2, and construction of mutant strains is described in detail below. GAS, GBS, and S. dysgalactiae subsp. equisimilis were routinely grown in Todd-Hewitt medium (BD Biosciences) supplemented with 2% (wt/vol) yeast extract (Amresco) (THY). For all reporter expression studies, bacteria were grown in chemically defined medium (CDM) (26) containing 1% (wt/vol) glucose at 37°C. When necessary, antibiotics were included at the following concentrations: chloramphenicol (Cm), 3 µg ml−1; erythromycin (Erm), 0.5 µg ml−1; spectinomycin (Spec), 100 µg ml−1. Cloning was performed using Escherichia coli strains DH10β (Invitrogen) and BH10C (47) grown in Luria-Bertani (LB) broth or on LB agar with antibiotics at the following concentrations: Erm, 500 µg ml−1; Spec, 100 µg ml−1.
TABLE 2 .
Bacterial strains used in this study
| Strain | Description | Source or reference(s) |
|---|---|---|
| NZ131 | Wild-type M49 S. pyogenes isolate | 48, 53 |
| JCC131 | NZ131 Δrgg3::cat; Cmr | 26 |
| BNL158 | NZ131 with integrated p7INT; Ermr | 54 |
| BNL170 | NZ131 shp2GGG shp3GGG; unmarked | This study |
| BNL177 | NZ131 shp2GGG shp3GGG with integrated pBL111; Ermr | This study |
| A909 | Wild-type S. agalactiae type Ia/C clinical isolate | 49, 55 |
| BLC100 | A909 ΔrovS; unmarked | This study |
| BLC101 | A909 shp1520GGG; unmarked | This study |
| BLC102 | A909 ΔrovS shp1520GGG; unmarked | This study |
| BLC103 | A909 with mutated putative RovS binding site; unmarked | This study |
| GGS-LT1 | S. dysgalactiae subsp. equisimilis strain isolated in Lin Tao laboratory and identified by 16S rRNA sequencing | This study |
Construction of plasmids for mutagenesis in GAS and GBS.
All plasmids and primers used in this study are described in Tables S1 and S2 in the supplemental material, respectively, in the supplemental material. For generation of a plasmid for shp2 start codon mutagenesis in GAS, a 1,115-bp fragment containing shp2 and flanking upstream and downstream regions was amplified by PCR using the primers BL80 and BL81 and cloned into the XmaI site of pFED760, generating pBL119. pBL119 was then used as the template for an inverse PCR reaction with the primer pair BL82/BL83 to generate a plasmid in which the shp2 translational initiation codon was mutated from ATG to GGG (pBL120). Separately, inverse PCR was performed using pLA101 as the template and primer pair JC139/JC140 to generate a plasmid in which the shp3 translational initiation codon was mutated to GGG (pJC180).
For generation of plasmids to mutate the shp1520 start codon or conserved region of the shp1520 promoter in GBS, a 2,252-bp fragment containing rovS, shp1520, and flanking upstream and downstream regions was amplified by PCR using the primers rovSregion-F1-notI/rovSregion-R1-notI and cloned into the NotI site of pFED760, generating pSar100. pSar100 was then used as the template for inverse PCR reactions with the primer pair gbsshp1520-mut-F1/gbsshp1520-mut-R1 or Pshp1520mut-F1/Pshp1520mut-R1 to generate plasmids in which the shp1520 start codon was mutated to GGG (pSar101) or two nucleotides in the conserved region of the shp1520 promoter were mutated (pSar107), respectively.
For generation of a plasmid to delete rovS, upstream and downstream DNA fragments flanking the rovS gene were amplified by PCR using the primer pairs rovSregion-F1-notI/rovS-dscomp-R1 and rovS-uscomp-F1/rovSregion-R1-notI. These products were purified and then fused in a second PCR reaction using the outside primers. This fusion product was cloned into the NotI site of pFED760 to generate pLCC1. To generate a plasmid for simultaneous deletion of rovS and mutation of the shp1520 start site, the rovS deletion plasmid pLCC1 was used as the template in an inverse PCR reaction with the primer pair gbsshp1520-mut-F1/gbsshp1520-mut-R1 to mutate the shp1520 start codon to GGG (pLCC2).
Generation of mutant GAS and GBS strains.
All GAS strains used in this study were derived from the serotype M49 strain NZ131 (48). All GBS strains used in this study were derived from the clinical isolate A909 (49). All deletion vectors were electroporated into NZ131 or A909, and a two-step temperature-dependent selection process was used to isolate mutants of interest (50). Briefly, cells containing each deletion construct were grown at the permissive temperature (30°C) and then shifted to 37°C and plated on the appropriate antibiotic to select for bacteria in which the plasmid had integrated at one of the flanking regions. Cells were then grown at the permissive temperature to allow the plasmid to recombine out of the chromosome, and loss of Erm resistance was used to identify a successful second crossover event and loss of the mutation vector. Genotypes were confirmed by PCR and sequencing. This process was repeated to construct double mutants.
Construction of luciferase transcriptional reporters.
Construction of the shp2 reporter plasmid, pBL111, was described previously (26). Site-specific integration of pBL111 at a tRNASer gene in BNL177 was confirmed by PCR. For a GBS reporter containing the shp1520 promoter region, 166 bp directly upstream of the shp1520 open reading frame were amplified using the primers Pshp1520-S1-bam/Pshp1520-lux-A1. Separately, the Vibrio fischeri luxAB genes were amplified from plasmid pCN59 (51) using the primers BL26/BL28. The shp1520 promoter product was fused in-frame to luxAB by overlap extension PCR using the primers Pshp1520-S1-bam/BL27. For generation of a shp1520 reporter that also included the entire upstream rovS gene, 1,097 bp directly upstream of the shp1520 open reading frame were amplified using the primers RovS-A1-bam/Pshp1520-lux-A1 and then fused to luxAB by overlap extension PCR using the primers RovS-A1-bam/BL27. The reporter fusion products were ligated into the BamHI and EcoRI sites of pLZ12Sp to generate pSar110 and pSar111. Reporter plasmids were electroporated into GBS and selected for using Spec resistance.
Luciferase transcriptional reporter assays
For luciferase assays, cells from overnight cultures grown at 30°C were diluted 100-fold into CDM and incubated at 37°C. At each time point, 50 µl of each culture was removed to an opaque 96-well plate, samples were exposed to decyl aldehyde (Sigma) fumes for 30 s, and luminescence (counts per second [CPS]) was quantified using a Turner Biosystems Veritas microplate luminometer. The optical density of the culture at 600 nm (OD600) was also measured at each time point using a Spectronic 20D spectrophotometer (Milton Roy). Relative light units were calculated by normalizing CPS to OD. For conditioned-medium experiments, donor strains were grown in CDM to an OD of 0.3 to 0.5, cells were spun down, and supernatants were passed through a 0.22-µm filter. Log-phase reporter cells [BNL177 or BLC101(pSar110)] were diluted 1:11 into conditioned supernatants, and light activity and OD were monitored as described above. In experiments containing synthetic peptides, reporter strains were grown in CDM to an OD of 0.3 to 0.5 and then diluted in fresh CDM containing 50 nM peptide of interest.
Coculture assays.
For coculture assays, reporter strains [BNL177 or BLC101(pSar110)] or donor strains [A909 and BLC101, or BNL158 and JCC131] were grown overnight at 30°C. GBS cells were diluted 100-fold into CDM, and GAS cells were diluted 75-fold to give approximately the same OD600 values. Equal volumes of donor and reporter cells were mixed together and incubated at 37°C, and 50 µl of each mixed culture was removed at various time points. Luminescence was measured as described above, and dilutions were then plated on THY plates with and without antibiotic to determine GAS and GBS CFUs.
Synthetic peptides.
Synthetic peptides were purchased from Neo-Peptide (Cambridge, MA). Purities of crude preparations used in luciferase assays ranged from 34% to 61%. Synthetic peptides were reconstituted as 2 mM stocks in dimethyl sulfoxide (DMSO) and stored at −80°C.
Biofilm assays.
Overnight cultures of GBS strains A909 and BLC101 were grown in THY at 30°C. Cultures were diluted 1:100 into CDM and grown at 37°C to an OD of 0.3 to 0.4 before cells were spun down, and supernatants were passed through a 0.22-µm filter. Overnight cultures of GAS strains BNL158 and BNL170 were diluted 1:50 into the conditioned supernatants from each strain, and 1 ml of these cultures was added to each well of a 24-well plate. A single autoclaved 1-cm-diameter circular aclar membrane (Honeywell Inc., Morristown, NJ) was added to each well, and the 24-well plate was incubated at 30°C for 24 h. Aclar membranes were then removed to a 6-well plate with 3 ml sterile water and rinsed for 2 to 3 min to remove nonadherent cells. Membranes were then removed to a 1.5-ml microcentrifuge tube with 1 ml phosphate-buffered saline (PBS) and sonicated for 3 min to removed attached cells. Tubes were spun down, and the bacterial pellet was resuspended in THY. Tenfold serial dilutions were plated on THY plates and incubated overnight at 37°C before CFUs were counted.
SUPPLEMENTAL MATERIAL
Plasmids used in this study
Primers used in this study
ACKNOWLEDGMENTS
Support for this work was provided by the NIH, grant AI091779, and the Burroughs Wellcome Fund, Investigators in the Pathogenesis of Infectious Disease Fellowship.
We are grateful to Grace Mattingly and Lin Tao, Donald Morrison, and Victor Nizet for providing us with bacterial strains used in these studies. Additional gratitude goes to Jennifer C. Chang for technical assistance and sharing of plasmids.
Footnotes
Citation Cook LC, LaSarre B, Federle MJ. 2013. Interspecies communication among commensal and pathogenic streptococci. mBio 4(4):e00382-13. doi:10.1128/mBio.00382-13.
ADDENDUM
Following review of the manuscript, an article by Fleuchot et al. was published showing the activity of Rgg/SHP pairs from three species of streptococcus (52). The findings are consistent with our observations.
REFERENCES
- 1. Ng W-L, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43:197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rasmussen TB, Givskov M. 2006. Quorum-sensing inhibitors as anti-pathogenic drugs. Int. J. Med. Microbiol. 296:149–161 [DOI] [PubMed] [Google Scholar]
- 3. LaSarre B, Federle MJ. 2013. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77:73–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Consortium THMP 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, Wade WG. 2010. The human oral microbiome. J. Bacteriol. 192:5002–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13:260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102:7952–7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beck JM, Young VB, Huffnagle GB. 2012. The microbiome of the lung. Transl. Res. 160:258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ, Active Bacterial Core Surveillance/Emerging Infections Program Network. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 299:2056–2065 [DOI] [PubMed] [Google Scholar]
- 10. Regan JA, Klebanoff MA, Nugent RP. 1991. The epidemiology of group B streptococcal colonization in pregnancy. Vaginal Infections and Prematurity Study Group. Obstet. Gynecol. 77:604–610 [PubMed] [Google Scholar]
- 11. Campbell JR, Hillier SL, Krohn MA, Ferrieri P, Zaleznik DF, Baker CJ. 2000. Group B streptococcal colonization and serotype-specific immunity in pregnant women at delivery. Obstet. Gynecol. 96:498–503 [DOI] [PubMed] [Google Scholar]
- 12. Barcaite E, Bartusevicius A, Tameliene R, Kliucinskas M, Maleckiene L, Nadisauskiene R. 2008. Prevalence of maternal group B streptococcal colonisation in European countries. Acta Obstet. Gynecol. Scand. 87:260–271 [DOI] [PubMed] [Google Scholar]
- 13. Verani JR, McGee L, Schrag SJ. 2010. Prevention of perinatal group B streptococcal disease—revised guidelines from CDC, 2010. MMWR Recomm. Rep. 59:1–36 [PubMed] [Google Scholar]
- 14. van der Mee-Marquet N, Fourny L, Arnault L, Domelier AS, Salloum M, Lartigue MF, Quentin R. 2008. Molecular characterization of human-colonizing Streptococcus agalactiae strains isolated from throat, skin, anal margin, and genital body sites. J. Clin. Microbiol. 46:2906–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cunningham MW. 2008. Pathogenesis of group A streptococcal infections and their sequelae. Adv. Exp. Med. Biol. 609:29–42 [DOI] [PubMed] [Google Scholar]
- 16. Roberts AL, Connolly KL, Kirse DJ, Evans AK, Poehling KA, Peters TR, Reid SD. 2012. Detection of group A Streptococcus in tonsils from pediatric patients reveals high rate of asymptomatic streptococcal carriage. BMC Pediatr. 12:3 http://dx.doi.org/doi:10.1186/1471-2431-12-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shaikh N, Leonard E, Martin JM. 2010. Prevalence of streptococcal pharyngitis and streptococcal carriage in children: a meta-analysis. Pediatrics 126:e557–e564. 10.1542/peds.2009-2648 [DOI] [PubMed] [Google Scholar]
- 18. Gupta R, Prakash K, Kapoor AK. 1992. Subclinical group A streptococcal throat infection in school children. Indian Pediatr. 29:1491–1494 [PubMed] [Google Scholar]
- 19. Dumre SP, Sapkota K, Adhikari N, Acharya D, Karki M, Bista S, Basanyat SR, Joshi SK. 2009. Asymptomatic throat carriage rate and antimicrobial resistance pattern of Streptococcus pyogenes in Nepalese school children. Kathmandu Univ. Med. J. (KUMJ) 7:392–396 [DOI] [PubMed] [Google Scholar]
- 20. Vijaya D, Sathish J, Jankiram K. 2013. The prevalence of group A streptococci carriers among asymptomatic school children. J. Clin. Diagn. Res. 7:446–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levy RM, Leyden JJ, Margolis DJ. 2005. Colonisation rates of Streptococcus pyogenes and Staphylococcus aureus in the oropharynx of a young adult population. Clin. Microbiol. Infect. 11:153–155 [DOI] [PubMed] [Google Scholar]
- 22. Gunnarsson RK, Holm SE, Söderström M. 1997. The prevalence of beta-haemolytic streptococci in throat specimens from healthy children and adults. Implications for the clinical value of throat cultures. Scand. J. Prim. Health Care 15:149–155 [DOI] [PubMed] [Google Scholar]
- 23. Mead PB, Winn WC. 2000. Vaginal-rectal colonization with group A streptococci in late pregnancy. Infect. Dis. Obstet. Gynecol. 8:217–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. 2011. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol. Microbiol. 80:1102–1119 [DOI] [PubMed] [Google Scholar]
- 25. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78:589–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 7:e1002190 http://dx.doi.org/doi:10.1371/journal.ppat.1002190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lasarre B, Aggarwal C, Federle MJ. 2013. Antagonistic Rgg regulators mediate quorum Sensing via competitive DNA binding in Streptococcus pyogenes. mBio 3(6):e00333-12 http://dx.doi.org/10.1128/mBio.00333-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ibrahim M, Nicolas P, Bessières P, Bolotin A, Monnet V, Gardan R. 2007. A genome-wide survey of short coding sequences in streptococci. Microbiology 153:3631–3644 [DOI] [PubMed] [Google Scholar]
- 29. Samen UM, Eikmanns BJ, Reinscheid DJ. 2006. The transcriptional regulator RovS controls the attachment of Streptococcus agalactiae to human epithelial cells and the expression of virulence genes. Infect. Immun. 74:5625–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mashburn-Warren L, Morrison DA, Federle MJ. 2012. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194:4589–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ibrahim M, Guillot A, Wessner F, Algaron F, Besset C, Courtin P, Gardan R, Monnet V. 2007. Control of the transcription of a short gene encoding a cyclic peptide in Streptococcus thermophilus: a new quorum-sensing system? J. Bacteriol. 189:8844–8854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carson HJ, Lapoint PG, Monif GR. 1997. Interrelationships within the bacterial flora of the female genital tract. Infect. Dis. Obstet. Gynecol. 5:303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rickard AH, Palmer RJ, Jr, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 60:1446–1456 [DOI] [PubMed] [Google Scholar]
- 35. Schauder S, Shokat K, Surette MG, Bassler BL. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463–476 [DOI] [PubMed] [Google Scholar]
- 36. Xavier KB, Bassler BL. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191–197 [DOI] [PubMed] [Google Scholar]
- 37. Wang BY, Kuramitsu HK. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 71:354–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stålhammar-Carlemalm M, Areschoug T, Larsson C, Lindahl G. 1999. The R28 protein of Streptococcus pyogenes is related to several group B streptococcal surface proteins, confers protective immunity and promotes binding to human epithelial cells. Mol. Microbiol. 33:208–219 [DOI] [PubMed] [Google Scholar]
- 39. Bröker G, Spellerberg B. 2004. Surface proteins of Streptococcus agalactiae and horizontal gene transfer. Int. J. Med. Microbiol. 294:169–175 [DOI] [PubMed] [Google Scholar]
- 40. Chmouryguina I, Suvorov A, Ferrieri P, Cleary PP. 1996. Conservation of the C5a peptidase genes in group A and B streptococci. Infect. Immun. 64:2387–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Franken C, Haase G, Brandt C, Weber-Heynemann J, Martin S, Lämmler C, Podbielski A, Lütticken R, Spellerberg B. 2001. Horizontal gene transfer and host specificity of beta-haemolytic streptococci: the role of a putative composite transposon containing scpB and lmb. Mol. Microbiol. 41:925–935 [DOI] [PubMed] [Google Scholar]
- 42. Davies MR, McMillan DJ, Beiko RG, Barroso V, Geffers R, Sriprakash KS, Chhatwal GS. 2007. Virulence profiling of Streptococcus dysgalactiae subspecies equisimilis isolated from infected humans reveals 2 distinct genetic lineages that do not segregate with their phenotypes or propensity to cause diseases. Clin. Infect. Dis. 44:1442–1454 [DOI] [PubMed] [Google Scholar]
- 43. Christensen KK, Christensen P, Flamholc L, Ripa T. 1974. Frequencies of streptococci of groups A, B, C, D, and G in urethra and cervix swab specimens from patients with suspected gonococcal infection. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 82:470–474 [DOI] [PubMed] [Google Scholar]
- 44. Forrer CB, Ellner PD. 1979. Distribution of hemolytic streptococci in respiratory specimens. J. Clin. Microbiol. 10:69–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Facklam R, Elliott J, Pigott N, Franklin AR. 1995. Identification of Streptococcus porcinus from human sources. J. Clin. Microbiol. 33:886–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87:1113–1132 [DOI] [PubMed] [Google Scholar]
- 47. Howell-Adams B, Seifert HS. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146–1158 [DOI] [PubMed] [Google Scholar]
- 48. McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, Scott J, Roe BA, Savic DJ. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190:7773–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lancefield RC, McCarty M, Everly WN. 1975. Multiple mouse-protective antibodies directed against group B streptococci. Special reference to antibodies effective against protein antigens. J. Exp. Med. 142:165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fleuchot B, Guillot A, Mézange C, Besset C, Chambellon E, Monnet V, Gardan R. 2013. Rgg-associated SHP signaling peptides mediate cross-talk in streptococci. PLoS One 8:e66042. 10.1371/journal.pone.0066042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Simon D, Ferretti JJ. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66:219–224 [DOI] [PubMed] [Google Scholar]
- 54. LaSarre B, Federle MJ. 2011. Regulation and consequence of serine catabolism in Streptococcus pyogenes. J. Bacteriol. 193:2002–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmids used in this study
Primers used in this study