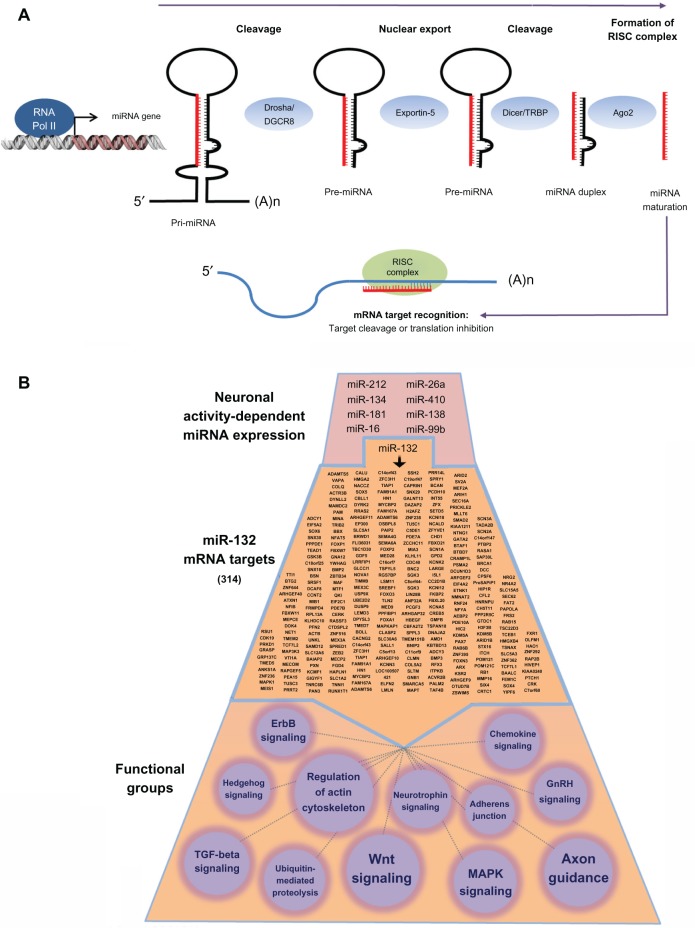
Figure 1.
Biogenesis and miRNA functionality in neurons.
Notes: (A) miRNA are transcribed from noncoding regions of the genome by RNA polymerase II, forming a hairpin loop (pri-miRNA) that is cleaved by Drosha/DGCR8 into a ~70 nt pre-miRNA. Pre-miRNA are exported from the nucleus in an exportin-5-dependent manner before further processing by Dicer. The mature strand of the miRNA is loaded into the RISC complex, where it binds to its target mRNA to inhibit translation. (B) Depiction of the complexity of miRNA functionality. At the top of the panel, we provide a limited list of brain-enriched miRNA that are inducibly expressed by neuronal activity. To gain an appreciation of the functional effects of a single miRNA, we provide a list of miR-132 mRNA targets (314 in total: middle section). This list was generated using the TargetScan algorithm. KEGG pathways analysis (bottom section) was used to generate functional classifications of the miR132 targets. Only a subset of the classifications is provided here. Circle size denotes the relative number of genes that make up the classification (the smallest functional class is Hedgehog signaling, which comprises six genes).
Abbreviations: GnRH, gonadotropin-releasing hormone; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; RNA, ribonucleic acid; mRNA, messenger RNA; miRNA, microRNA; nt, nucleotide; RISC, RNA-induced silencing complex; TGF, transforming growth factor; RNA pol II, RNA polymerase II; RISC, RNA-induced silencing complex; TAR, trans-activation response; TRBP, TAR RNA binding protein.