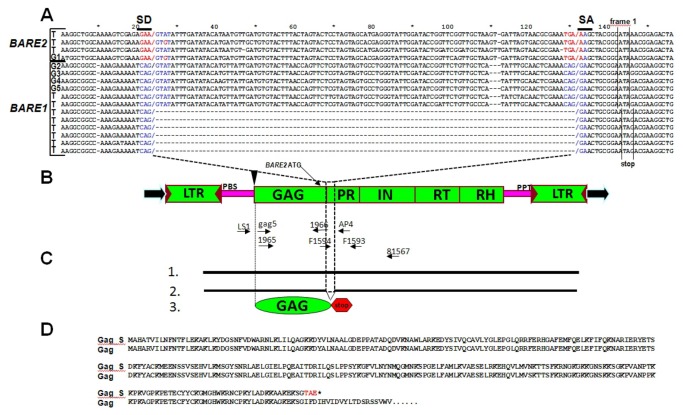
Figure 2. Splicing of BARE1.
A. Alignment of a set of BARE genomic DNA sequences and cDNA clones showing the forms with the consensus splice junctions (SD and SA) within the gag domain. Nucleotides shaded blue match the consensus slice junctions, those in red do not. Genomic DNA sequences are labelled as “G”, cDNA as “T”. Genomic sequences in the alignment are: G1, AJ279072; G2, Z17327; G3, AY66155; G4, AY485643; G5, BQ900685. B. Schematic diagram of BARE retrotransposons showing the LTRs, encoded proteins of the open reading frame, and the cDNA priming sites (PBS, PPT), together with the position of the diagnostic primers as arrows below. The inverted triangle indicates the 8 bp deletion of the start codon in BARE2 that eliminates synthesis of Gag; the following ATG for the Pol domain is indicated. C. Diagrams of the unspliced (1) and spliced (2) forms of the BARE1 transcripts as well as the translated product of the spliced form (3). D. Conceptual translation of the BARE1 ORF (Accession Z17327) covering the Gag and part of the PR region for the unspliced (Gag) and spliced (Gag_S) transcript forms. Amino acids altered by the splice-induced frameshift are shown in red, the stop codon as *.