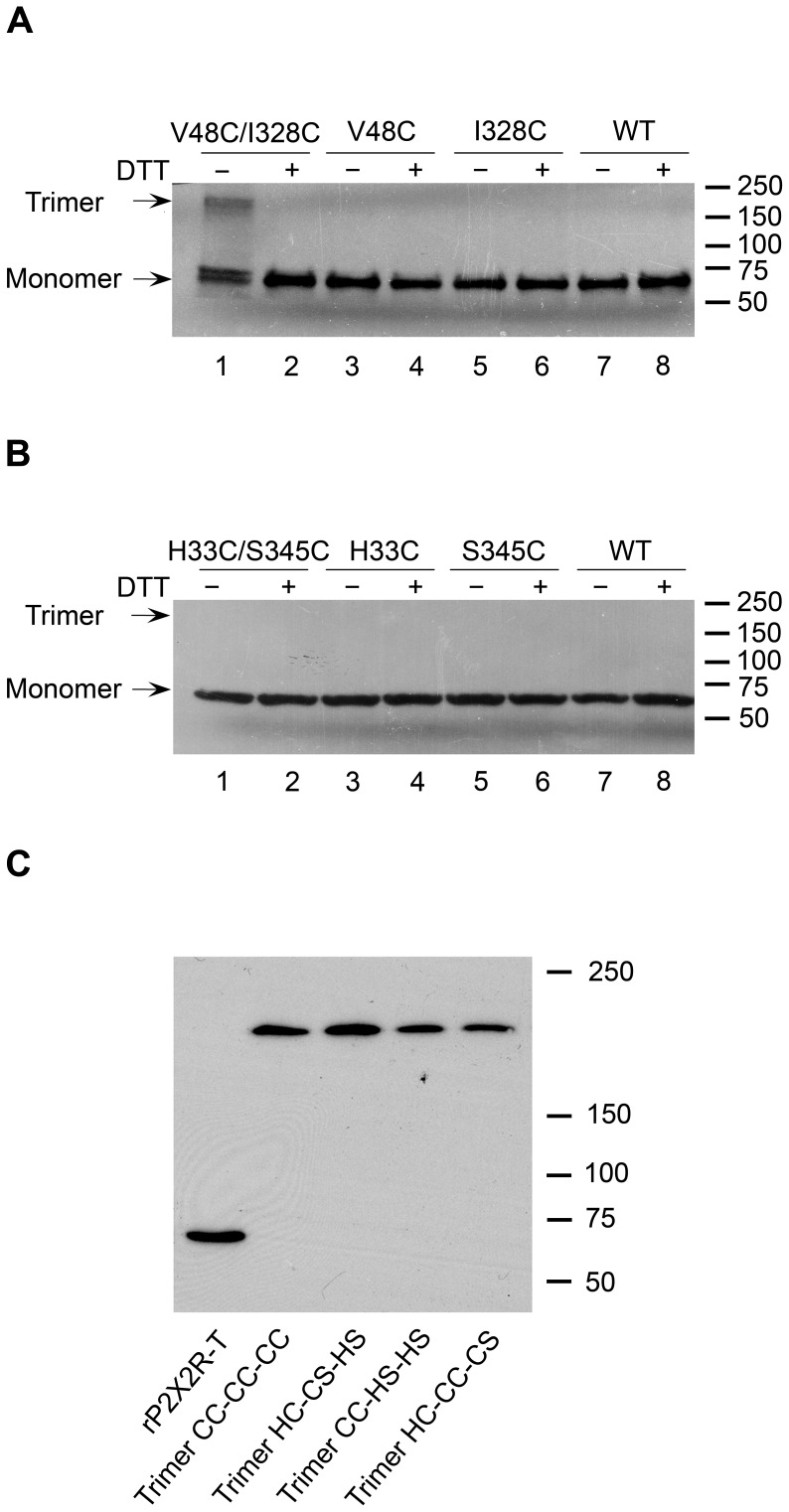
Figure 3. Western blot analysis.
(A) Inter-subunit disulfide bond formation between V48C and I328C in the rP2X2R. Double mutant V48C/I328C, single mutants V48C and I328C and wild-type rP2X2R were transiently expressed in HEK293 cells. Protein samples were extracted from the membrane. (B) Analysis of specific trimer formation in double mutant H33C/S345C, single mutants H33C and S345C and wild-type rP2X2R. In (A) and (B), all the single mutants and the wild type protein served as negative controls to estimate the background of nonspecific disulfide bond formation. Arrows indicate monomers and trimers. Above lanes 2, 4, 6, and 8 in (A) and (B), “+” means protein samples were loaded with DTT to denature the disulfide bond. Above lanes 1, 3, 5, 7 in (A) and (B), “–” means protein samples were loaded without DTT. Proteins were separated on SDS-PAGE gels (8%) and detected by Western blotting via a FLAG-tag antibody. Protein molecular weight markers (kDa) are indicated on the right. These results were observed in at least four independent experiments for each receptor. (C) Western blot analysis of the concatamerised trimers. The rP2X2R-T monomer, trimers CC-CC-CC, CC-HS-HS, HC-CS-HS, and HC-CC-CS were transiently expressed in HEK293 cells. H and S mean His33 and Ser345, respectively. C means cysteine substitution. In the monomer, each subunit has one N terminus and one C terminus. The concatameric trimer constructs have only one N terminus and one C terminus. Subunit organizations of concatameric trimer constructs are presented in Figure 4A. Protein samples were extracted from the membrane, separated by SDS-PAGE gels (8%) under reducing conditions, and detected by Western blotting with rP2X2 antibody. The positions of molecular mass standards (kDa) are shown on the right. The trimers revealed a single band indicating the same size (∼186 kDa) and remained intact. These results were observed in at least four independent experiments for each receptor.