Abstract
Modifications to DNA and histone tails represent key epigenetic marks involved in establishing and maintaining cell identity and can be dysregulated in human diseases, including cancer. Two such modifications, tri-methylation of lysine-27 on histone H3 (H3K27me3) mediated by the Polycomb complex and hydroxymethylation of cytosines on DNA, have recently been shown to be dynamically regulated during differentiation. Here, we show that global levels of 5-hydroxymethylcytosine (5hmC) and H3K27me3 are highly correlated across a variety of somatic tissues. In multiple hierarchically organized tissues, both marks showed almost identical cell-by-cell distribution patterns that exhibited a tight association with differentiation. In particular, tissue stem cell compartments were characterized by low levels of both marks, whereas differentiated cell compartments exhibited high levels of 5hmC and H3K27me3. This pattern of correlation between the two marks could be recapitulated in an in vitro model system of induced differentiation in prostate epithelial cells. While the correlation between 5hmC and H3K27me3 levels is also maintained in human cancers, the degree of correlation is reduced. These findings suggest a previously unappreciated link between 5hmC and H3K27me3 regulation that should be explored in future mechanistic studies.
Keywords: 5-hydroxymethylcytosine, DNA methylation, differentiation, cancer, H3K27me3, polycomb, epigenetics
Introduction
Epigenetic modifications comprise a series of heritable changes in chromatin organization that do not affect the primary DNA sequence. Two of the best-studied classes of epigenetic marks are (1) modifications of the DNA molecule itself, such as methylation of the 5-position of cytosines (5 mC); and (2) posttranslational modifications of histone tails in nucleosomes. The complex interactions between such DNA methylation marks and histone modifications can regulate genome organization and can orchestrate tissue-specific gene expression patterns during development and differentiation.1 Importantly, many disease states are characterized by alterations of epigenetic marks on DNA and histones; these alterations are often associated with aberrant genome organization and gene expression. Specifically, human cancers almost universally develop dysregulation of epigenetic marks, during both cancer initiation and disease progression.2
DNA methylation patterns in human health and disease have been extensively studied over the past four decades.3,4 In particular, it has recently been shown that methylated cytosines (5mC) in DNA can be further oxidized to 5-hydroxymethylcytosine (5hmC). As a modified base, 5hmC has been detected in mammalian and viral genomes as early as the 1950s using crude fractionation methods.5 However, the recent discovery that members of the ten-eleven translocated protein family (TET1, TET2, TET3) can enzymatically convert 5mC to 5hmC in a targeted manner has spurred a renewed interest in this DNA modification.6-8 Previous studies have suggested that 5hmC plays a crucial role in stem cell biology and lineage-specific differentiation.9,10 In an attempt to further elucidate the biological function of 5hmC, several groups developed novel methodologies to investigate the genome-wide distribution pattern of 5hmC in embryonic stem cells.11-15 Taken together, these reports showed that 5hmC can be found around the transcriptional start sites of active and repressed gene promoters, suggesting that 5hmC could be involved in transcriptional regulation. Furthermore, 5hmC was enriched at genes whose promoters bear “bivalent” histone 3 lysine 27 trimethylation (H3K27me3) repression marks and histone 3 lysine 4 trimethylation (H3K4me3) activation marks. This “bivalent” chromatin state is thought to poise gene loci for further epigenetic regulation, suggesting that 5hmC could be involved in the transcriptional regulation and fine-tuning of transcriptional output during ES cell differentiation.
More generally, histone H3 lysine 27 trimethylation (H3K27me3) is a well-known histone modification regulated during ES cell differentiation and development. H3K27me3 levels are thought to be regulated by the histone methyltransferase enhancer of zeste 2 (EZH2), as well as the lysine demethylases JMJD3 and UTX.16 EZH2 is the catalytic subunit of the polycomb repressive complex 2 (PRC2), and trimethylation of H3K27 by PRC2 recruits the PRC1 complex, resulting in gene silencing during ES cell differentiation.17 H3K27me3 is a polycomb mark usually associated with heterochromatin, and transcriptional repression and is known to be dysregulated in cancer.18,19
We have recently demonstrated that global levels of 5hmC are high in terminally differentiated cells in most adult tissues,20 suggesting a key role of 5hmC in tissue-specific differentiation. Conversely, the stem cell compartments in hierarchically differentiated tissues show greatly reduced 5hmC levels compared with their more differentiated counterparts. This distinct distribution pattern with a clear enrichment for terminally differentiated cells is highly reminiscent of the distribution recently described for H3K27me3.21 Here we show that 5hmC and H3K27me3 are tightly correlated on a cell-by-cell basis in multiple normal human tissues. Although to a somewhat reduced extent, this association is maintained in human cancers, where both marks are significantly downregulated at a global level. These observations suggest a previously unappreciated link between 5hmC and H3K27me3.
Results and Discussion
Global levels and cellular localization of 5hmC and H3K27me3 are highly correlated
We previously showed that 5hmC global content and global levels of H3K27me3 each track with differentiation in multiple normal tissues in two separate studies.21 We therefore hypothesized that these marks may be directly correlated with each other in stem and differentiated cell compartments. We began to explore this hypothesis by examining the levels of 5hmC and H3K27me3 in colonic mucosa, which represents a prototypical example of a hierarchically organized epithelial tissue. In the hierarchically organized colonic epithelium, the crypts harbor the stem cell compartment where colonocytes are formed by asymmetric division.22 Conversely, mature and terminally differentiated colonocytes are localized on the luminal side of the colon. We used previously validated immunohistochemical approaches developed by our group to evaluate the global levels of 5hmC and H3K27me3 in directly adjacent tissue sections from the human colon.20,21 Interestingly, we observed a high degree of concordance between 5hmC and H3K27me3 levels, in which both marks were observed at high levels in terminally differentiated apical cells, whereas cells at the base of the colonic crypts showed greatly reduced staining of both marks (Fig. 1). This observation provided initial evidence for a potential co-regulation of 5hmC and H3K27me3 during differentiation.
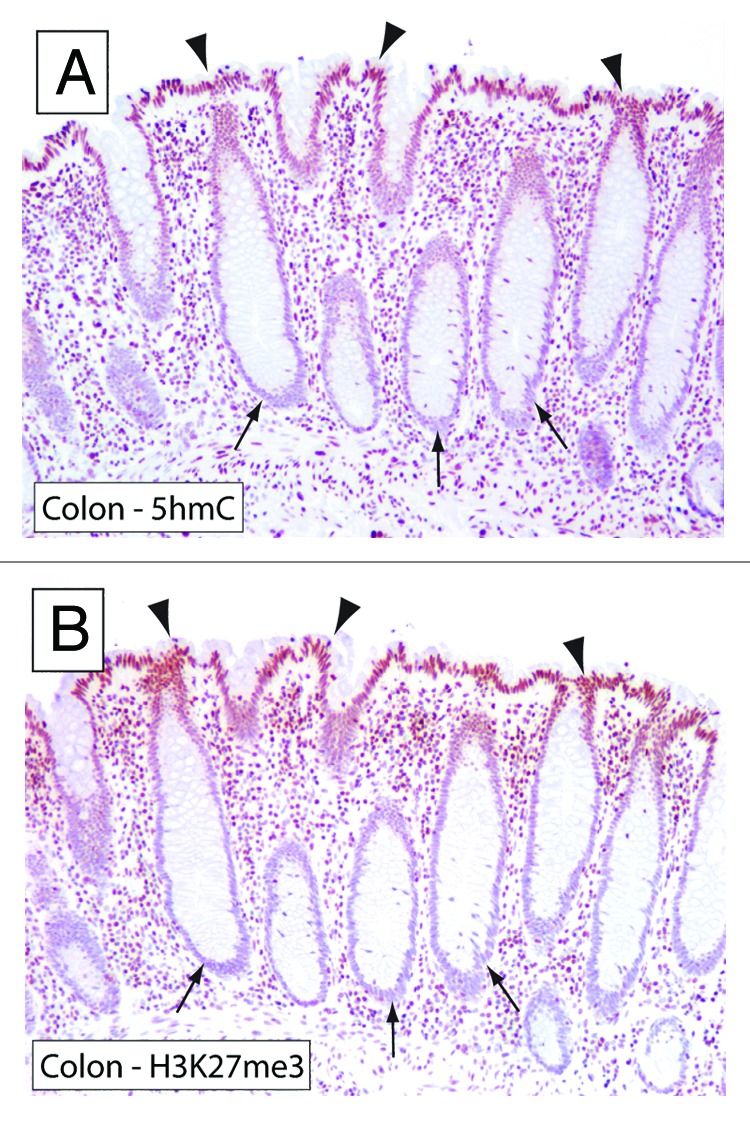
Figure 1. 5hmC and H3K27me3 show highly similar distribution patterns in colonic mucosa. To evaluate the distribution of 5hmC and H3K27me3 in the colonic mucosa, adjacent sections of formalin-fixed paraffin-embedded colonic tissues were stained with 5hmC and H3K27me3-specific antibodies. Note that terminally differentiated epithelial cells toward the lumen of the colon (arrowheads) show strong staining for 5hmC and H3K27me3, whereas cells in the crypts (arrows) show very low levels of 5hmC and H3K27me3.
To further evaluate the degree of co-localization of 5hmC and H3K27me3 on a cell-by-cell level, we used double-label immunofluorescence microscopy to detect both marks in the same tissue section.
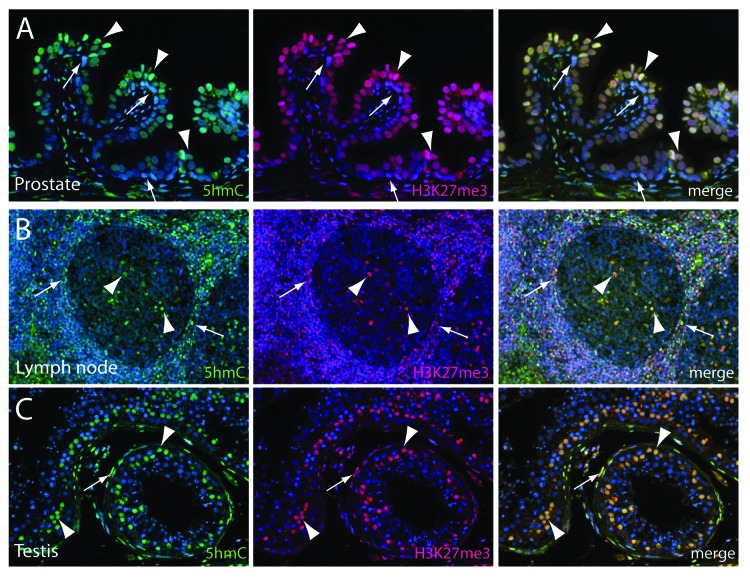
Secretory glands of the adult prostate are lined by a two-layer epithelium comprised of a luminal (Fig. 2A, arrowheads) and a basal (Fig. 2A, arrows) cell compartment. Similar to the colonic mucosa, the basal cell layer harbors the regenerative stem-like compartment, whereas the luminal cells are thought to be terminally differentiated.23 As shown previously, 5hmC was abundant in the luminal cell compartment but greatly reduced in the basal cell compartment (Fig. 2A).20 Similarly, global H3K27me3 levels were much higher in luminal cells as compared with basal cells of the prostate, suggesting a differentiation-dependent co-regulation of both marks in the prostatic epithelium. It is worth noting that the staining levels for 5hmC showed a slight cell-to-cell variability within the luminal cell compartment, with some cells showing stronger staining intensities than others (Fig. 2A). Remarkably, these different staining levels were also reflected in the H3K27me3 staining, indicating a tight correlation of 5hmC and H3K27me3, even at the level of individual cells.
Figure 2. Immunofluorescence double labeling reveals a high degree of co-localization of 5hmC and H3K27me3 in differentiated adult tissues. Tissue sections containing normal prostate epithelium, lymphoid tissue and testis were co-immunolabeled with antibodies specific to 5hmC (shown in green) and H3K27me3 (shown in red). (A) In normal prostate epithelium, 5hmC and H3K27me3 are present at high levels in the terminally differentiated luminal cells (arrowheads). Note that basal cells (arrows) exhibit greatly decreased staining intensities for both marks. (B) In the activated lymph follicle, 5hmC and H3K27me3 are both present with very high concordance in distinct cell populations in the germinal center (arrowheads) and the marginal zone (arrows). (C) In cross sections of the human testis, 5hmC and H3K27me3 are present in Sertoli cells in the seminal tubules (arrowheads) and in stromal cells surrounding the tubuli (arrows), but not in spermatogonia or mature spermatids.
As part of the immune system, lymph follicles are highly organized structures with distinct cellular organization patterns, which enable B-cell proliferation and maturation. The germinal center (Fig. 2B, arrowheads), which shows a high density of proliferating B-lymphocytes, is surrounded by the paracortex, which mainly consists of T-cells. Interestingly, the staining distribution of 5hmC and H3K27me3 in such secondary lymph follicles showed a very distinct pattern. The marginal zone (Fig. 2B, arrows) of the follicle exhibited a large number of cells with high 5hmC and H3K27me3 levels, whereas cells in the germinal center were mostly negative for both marks (Fig. 2B). Only a subset of cells in the germinal center showed high levels of 5hmC and H3K27me3 (Fig. 2B, arrowheads). This distinct distribution pattern could suggest that proliferating and maturing B-cells are characterized by low levels of 5hmC and H3K27me3.
Furthermore, in human testis, 5hmC and H3K27me3 showed an almost identical staining pattern (Fig. 2C). Both marks were present at high levels in terminally differentiated Sertoli cells within the tubuli seminiferi and in stromal cells surrounding the tubuli. Spermatogonia and mature spermatids, however, were devoid of 5hmC and H3K27me3 staining. In each of these systems, prostate, lymphoid tissue and testis, 5hmC and H3K27me3 levels were highly correlated even at a cell-by-cell level.
5hmC and H3k27me3 levels increase during induced differentiation in vitro
To recapitulate the changes in global 5hmC and H3K27me3 levels in a model system of induced differentiation, we used the normal prostate epithelial line RWPE-1.24 RWPE-1 cells show a basal like prostate epithelium phenotype characterized by low levels of 5hmC, H3K27me3 and absence/very low expression of the androgen receptor (AR) when cultured under standard cell culture conditions (2D culture) (Fig. 3). However, when cultured in a 3D matrix containing laminin and collagen, RWPE-1 cells showed robust glandular differentiation characterized by the formation of branching acini.25-27 This induced acinar differentiation was associated with an increase of luminal differentiation specific markers such as AR (Fig. 3B). Strikingly, 5hmC and H3K27me3 levels were also greatly increased in the acinar structures generated in the 3D culture system, despite being almost undetectable under conventional 2D culture conditions. Therefore this model system allows a partial recapitulation of prostatic epithelial differentiation and highlights the tight association of 5hmC and H3K27me3. Along these lines, it is worth noting that induced differentiation in other model systems has been shown to lead to an accumulation of 5hmC and reduction of 5mC specifically in enhancer and promoter regions, a step that might be required for establishing differentiation specific chromatin states.28,29
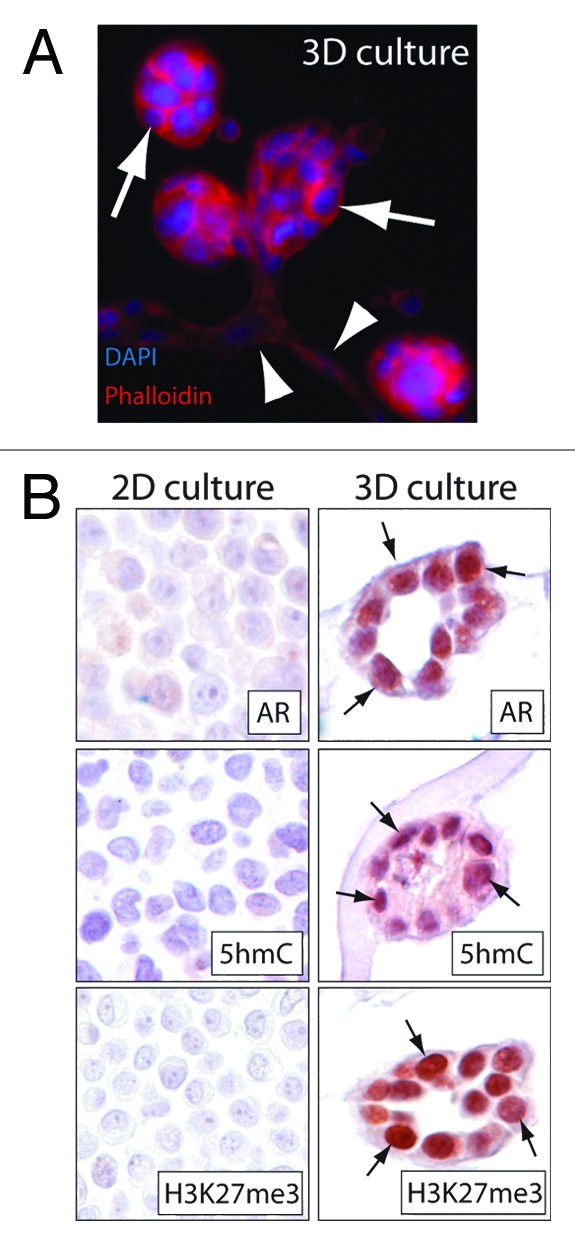
Figure 3. In vitro differentiation of prostate epithelial cells is associated with increased 5hmC and H3K27me3 levels. RWPE-1 prostate epithelial cells showed a basal cell like phenotype characterized by the absence of androgen receptor (AR) expression and low to undetectable levels of 5hmC and H3K27me3 when cultured under standard 2D culturing conditions. (A) Introduction of RWPE-1 cells in a 3D matrigel matrix induced formation of highly organized acinar structures (arrows) and ductal branching (arrowheads). This morphological differentiation was associated with an increase in AR levels and accumulation of 5hmC and H3K27me3 (arrows). (B) shows representative micrographs of cross sections obtained from 2D cultures and 3D cultures. Note the high level of AR, 5hmC and H3K27me3 staining in the acinar epithelial structures formed in 3D cultures (arrows).
Global 5hmC and H3K27me3 levels are reduced in solid tumors
Tumor initiation and progression are associated with a wide range of genetic and epigenetic changes.2,30,31 Changes in DNA methylation patterns are almost universal in human cancers and can be found at early stages during tumor progression. The dysregulation of DNA methylation patterns in cancers often involves both global DNA hypomethylation and gain of methylation marks in CpG islands.2 We have recently provided the first evidence that global 5hmC levels are greatly reduced in multiple human carcinomas compared with normal tissue.20 This finding was independently corroborated by a number of groups and expanded to other tumor types, suggesting that the bulk of the neoplastic cells in solid tumors including carcinoma of the breast, prostate, colon, malignant glioma and melanoma show profound decreases in 5hmC levels when compared with their differentiated normal counterparts.20,32-36 Moreover the loss of 5hmC is far more pronounced than the decrease in 5mC levels observed in many tumor types, suggesting that the reduction in 5hmC occurs independently of reductions in 5mC.20 This reduction of 5hmC levels appears to be an early event; in some tumors, it occurs at the stage of pre-invasive lesions (Haffner et al. unpublished data). Furthermore, recent evidence suggests that the extent of 5hmC loss in tumor cells tracks with histopathological grade and appears to be a prognostic indicator in glioblastoma multiforme and melanoma.34,36 The mechanism for this profound loss of 5hmC is currently under investigation, and several hypotheses have been proposed. For instance, in 30% of myeloid malignancies, TET2 appears to be inactivated by deletion or somatic mutations.37,38 This genetic inactivation is associated with a decrease in 5hmC levels.35 Consistent with a “driver” role for these mutations, Tet2-deficient mice show an enlargement of the hematopoietic stem cell compartment and develop myeloproliferative disorders.39 In solid tumors, however, mutations in TET genes are observed less frequently and are therefore not likely to contribute to the almost universally observed decrease of 5hmC. However, loss of 5hmC is frequently accompanied by reduced mRNA expression of TET1, TET2 and TET3 in a variety of solid tumors.32 Moreover, recent in vivo data also demonstrate that loss of TET1 and TET2 results in increased tumor growth and invasion and a global reduction in 5hmC, suggesting that TET1 and TET2 could function as tumor suppressor genes.36,40 Furthermore, metabolic alterations, such as the generation of TET-inhibitory metabolites like 2-hydroxyglutarate through mutant IDH1 and IDH2, have been recently discussed as potential causes for TET enzyme dysfunction and consequently 5hmC loss in tumors.41,42
The role of 5hmC in epigenetic regulation appears to be regulated by a complex network of enzymes.8,43,44 For instance, recent evidence suggests that 5hmC is likely to be an intermediate in an active de-methylation process in which the first step involves the oxidation of 5mC to 5hmC. It was postulated that 5hmC could get further oxidized to 5-formylcytosine (5fC) or 5-carboxylcytosine (5caC) in a process that involves TET enzymes.9,43-48 5fC and 5caC can then get excised by thymine-DNA glycosylase TDG and base excision repair.47,49 Alternatively, it has been proposed recently that 5hmC could be deaminated by the DNA methyltransferases DNMT3a and DNMT3b and then further repaired in a process involving DNA-glycosylases and base excision repair.50 The complexity of 5hmC turnover and the potential dynamics of this process present multiple pathways that, if corrupted, could lead to reduced 5hmC levels in tumors.
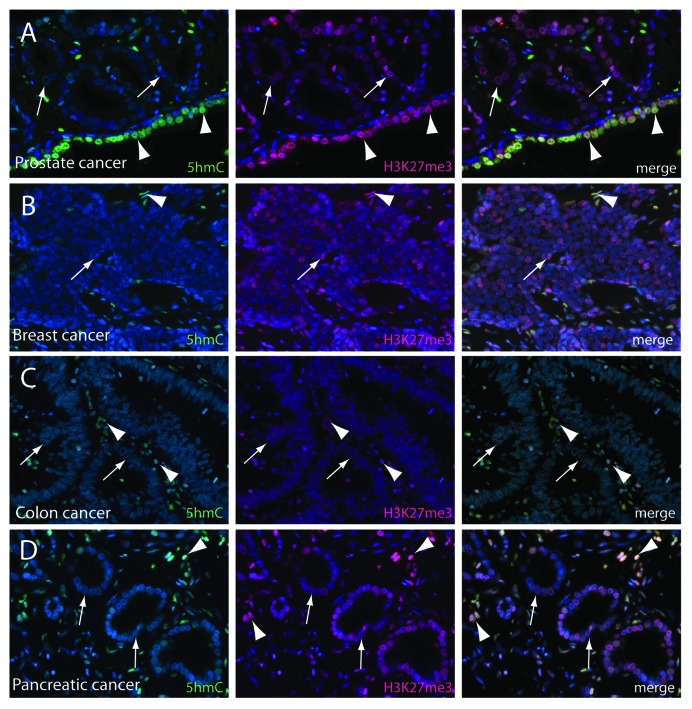
Similar to 5hmC, global H3K27me3 levels have also been shown to be reduced in breast, ovarian, pancreatic and prostate cancer and low levels of the mark have been associated with worse prognosis.18,21 Given the tight co-regulation of 5hmC and H3K27me3 in normal tissue, we aimed to further investigate these two epigenetic marks in prostate, breast, colon and pancreatic cancer (Fig. 4). In line with previous reports, we observed a profound loss of 5hmC in neoplastic cells, whereas adjacent normal epithelial or stroma cells showed robust staining. H3K27me3 levels appeared generally reduced in cancer samples; however, at close scrutiny, the loss H3K27me3 appeared overall much less pronounced. This was particularly evident in lower-grade lesions (Fig. 4). Furthermore, we also observed that single cells within the tumor that showed no detectable 5hmC levels showed strong staining for H3K27me3, suggesting at least a partial uncoupling of the tight correlation of these two marks in cancer cells compared with normal tissues.
Figure 4. 5hmC and H3K27me3 levels are decreased in cancers. (A) Prostate adenocarcinoma (arrows) showed global decreased levels of 5hmC and H3K27me3 as compared to normal prostate luminal cells (arrowheads). Similarly, neoplastic cells in breast (B), colon (C) and pancreatic adenocarcinoma (D) (arrows) were characterized by reduced 5hmC and H3K27me3 staining levels. Tumor associated stromal cells (arrowheads) showed high levels of 5hmC and H3K27me3. Note that the degree of loss between adjacent normal tissue and cancer cells was more pronounced for 5hmC and the correlation of 5hmC and H3K27me3 is less pronounced.
Interestingly, high levels of 5hmC and H3K27me3 can for the most part only be found in quiescent, non-proliferating cells. Replicating cells, however, show low levels of 5hmC and H3K27me3, suggesting that both marks are not actively maintained during replication. This is in agreement with a recent report showing the passive, replication-dependent loss of 5hmC in murine pre-implantation blastomeres.51 It remains to be shown if passive replication-dependent loss can also explain the 5hmC and H3K27me3 distribution patterns observed in normal adult tissues and the alteration of these patterns in cancers.
In conclusion, we show that global levels of 5hmC and H3K27me3 are tightly co-regulated during hierarchical differentiation in adult tissues. Furthermore, we show that in solid tumors both marks are decreased. Overall these findings suggest that 5hmC and H3K27me3 are linked by a yet unidentified mechanism.
Materials and Methods
Formalin-fixed paraffin-embedded tissue sections were de-paraffinized and then steamed for 40 min in EDTA solution (Zymed) followed by 5 min incubation in 3.5 N HCl. Immunolabeling was performed with rabbit polyclonal anti-5hmC antibodies (1:20K dilution, Active Motif) for 45 min or mouse monoclonal anti-H3K27me3 antibodies (1:75 dilution, ab6002, Abcam) overnight at 4°C as described previously.21 For detection of androgen receptor (AR), slides were steamed in HTTR buffer for 50 min and incubated with anti-AR rabbit polyclonal antibodies (1:1,000 dilution, N20, Santa Cruz Biotechnologies) for 45 min. Immune complexes were then visualized using the Power Vision+ poly-HRP IHC Kit (ImmunoVision Inc) using 3,3′-diaminobenzidine (Sigma) as a chromogen.
For co-immunolabeling of 5hmC and H3K27me3, antibodies were used at 1:6,000 and 1:75 dilutions, respectively. Immune complexes were visualized with Alexa 488 nm anti-rabbit and Alexa 565 nm anti-mouse (Life Technologies) secondary antibodies. After nuclear counterstaining with DAPI, slides were coverslipped with Prolong (Life Technologies). All slides were imaged on a Nikon 50i epifluorescence microscope. Immunofluorescence images were captured using a CoolsnapEZ digital camera (Photometrics) and the Nikon NIS-Elements (Nikon) software package.
RWPE-1 cells were cultured in KSFM containing 0.2 ng/ml epidermal growth factor (EGF) and 25 mg/ml bovine pituitary extract (Invitrogen). 3D culture experiments were performed as described previously27,52 and were either stained directly in situ with rhodamine phalloidin (Life Technologies) or embedded in paraffin and processed for immunohistochemistry as described above.
Acknowledgments
The authors would like to acknowledge Marcella Sutherland and Bonnie Gambichler from the Johns Hopkins TMA core facility and the Brady Urological Institute Prostate Specimen Repository for providing and processing tissue specimens and Dr Alan K. Meeker for helpful discussion. This study was supported in part by grants from the National Institutes of Health/National Cancer Institute (CA58236, CA070196 P50CA58236), the Prostate Cancer Foundation, the Patrick C. Walsh Prostate Cancer Research Fund, a generous gift from David H. Koch (to S.Y., W.G.N. and A.M.D.), the V Foundation for Cancer Research Martin D. Abeloff V Scholar Award (S.Y.) and the Cleveland Foundation Ellen B. Masenhimer Fellowship (S.Y.). M.C.H. is supported by a Young Investigator Award from the Prostate Cancer Foundation. A.M.D. is currently employed at Predictive Biosciences, Inc as well being part-time adjunct Professor of Pathology, Oncology and Urology at the Johns Hopkins University School of Medicine. No funding or other support was provided by Predictive Biosciences, Inc for any of the work in this manuscript.
Disclosure of Potential Conflicts of Interest
The terms of the relationship between A.M.D. and Predictive Biosciences, Inc are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies. No potential conflicts of interest were disclosed by any other authors.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25010
References
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 4.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyatt GR, Cohen SS. The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine. Biochem J. 1953;55:774–82. doi: 10.1042/bj0550774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 9.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–13. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 12.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–7. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Zhang Y. Tet1 and 5-hydroxymethylation: a genome-wide view in mouse embryonic stem cells. Cell Cycle. 2011;10:2428–36. doi: 10.4161/cc.10.15.16930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10:669–82. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y, Xia W, Zhang Z, Liu J, Wang H, Adsay NV, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol Carcinog. 2008;47:701–6. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613–8. doi: 10.1158/1078-0432.CCR-10-2156. [DOI] [PubMed] [Google Scholar]
- 20.Haffner MC, Chaux A, Meeker AK, Esopi DM, Gerber J, Pellakuru LG, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–37. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellakuru LG, Iwata T, Gurel B, Schultz D, Hicks J, Bethel C, et al. Global levels of H3K27me3 track with differentiation in vivo and are deregulated by MYC in prostate cancer. Am J Pathol. 2012;181:560–9. doi: 10.1016/j.ajpath.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 23.De Marzo AM, Nelson WG, Bieberich CJ, Yegnasubramanian S. Prostate cancer: New answers prompt new questions regarding cell of origin. Nat Rev Urol. 2010;7:650–2. doi: 10.1038/nrurol.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bello-DeOcampo D, Kleinman HK, Deocampo ND, Webber MM. Laminin-1 and alpha6beta1 integrin regulate acinar morphogenesis of normal and malignant human prostate epithelial cells. Prostate. 2001;46:142–53. doi: 10.1002/1097-0045(20010201)46:2<142::AID-PROS1018>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Webber MM, Bello D, Kleinman HK, Hoffman MP. Acinar differentiation by non-malignant immortalized human prostatic epithelial cells and its loss by malignant cells. Carcinogenesis. 1997;18:1225–31. doi: 10.1093/carcin/18.6.1225. [DOI] [PubMed] [Google Scholar]
- 26.Härmä V, Virtanen J, Mäkelä R, Happonen A, Mpindi JP, Knuuttila M, et al. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One. 2010;5:e10431. doi: 10.1371/journal.pone.0010431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyson DR, Inokuchi J, Tsunoda T, Lau A, Ornstein DK. Culture requirements of prostatic epithelial cell lines for acinar morphogenesis and lumen formation in vitro: role of extracellular calcium. Prostate. 2007;67:1601–13. doi: 10.1002/pros.20628. [DOI] [PubMed] [Google Scholar]
- 28.Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–65. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, et al. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat Commun. 2012;3:818. doi: 10.1038/ncomms1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–9. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–5. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orr BA, Haffner MC, Nelson WG, Yegnasubramanian S, Eberhart CG. Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One. 2012;7:e41036. doi: 10.1371/journal.pone.0041036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–43. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lian CG, Xu Y, Ceol C, Wu F, Larson A, Dresser K, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135–46. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–7. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–79. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- 41.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo JU, Su Y, Zhong C, Ming GL, Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–8. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J Biol Chem. 2011;286:35334–8. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J Biol Chem. 2012;287:33116–21. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]