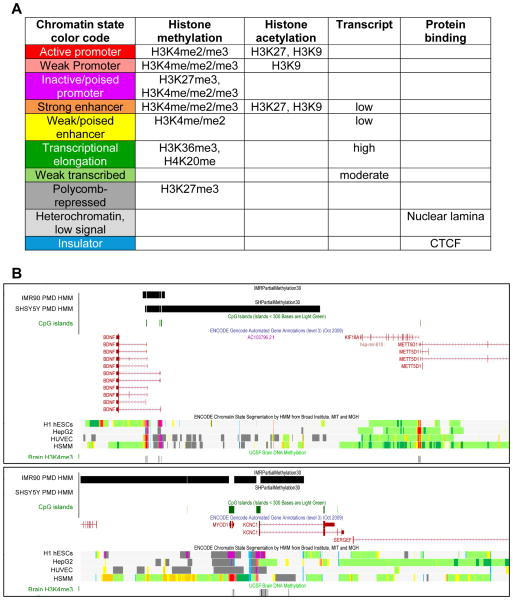
Figure 2. Chromatin states defined by histone modifications and protein binding.
A. A color-coded guide to the chromatin state maps derived from hidden Markov model (HMM) segmentation of histone modifications and CTCF binding sites in non-neuronal human cell lines from the human ENCODE project [36]. Histone methylation and acetylation was analyzed by ChIP-seq using antibodies to the specific modifications and sites listed. Transcript analysis was determined by RNA-seq on the same cell lines. Genes involved in developmental and tissue-specific functions exhibit H3K27 trimethylation (H3K27me3) blocks characteristic of polycomb-repressed genes (dark grey) and have inactive but “poised” bivalent promoter states (purple) of both silent (H3K27me3) and active (H3K4me3) histone modifications in human embryonic stem cells (hESCs).
B. Two examples of chromatin state maps combined with PMD state maps in the UCSC Genome Browser for neurologically relevant genes. PMDs mapped from HMM analysis of MethylC-seq data in fibroblast (IMR90) or neuronal (SH-SY5Y) cell lines [3] are shown as black bars, with gaps at CpG islands (green) because they were removed from the PMD analysis. ENCODE tracks (middle) include annotated genes and chromatin state segmentation by HMM color coded as in A. While neuronal tissue was not included in the preliminary ENCODE analysis of chromatin states, a track of “Brain H3K4me3” is shown at the bottom that identifies active genes in human adult brain. Notice the difference in chromatin states between regions covered by tissue-specific PMDs (BDNF, MYOD1, KCNC1, inactive poised promoter, polycomb-repressed, and heterochromatin) compared to genes with HMD in all tissues (METT5D1, KF18A, active promoter and transcribed). Also, notice the difference in chromatin states at the MYOD1 locus in human skeletal muscle myotubule (HSMM) cells compared to embryonic stem cells (H1 hESCs, inactive/poised promoter) and liver (HepG2) or umbilical vein epithelial cells (HUVEC) which have the H3K27me3 marks of polycomb repression. Both MYOD1 and KCNC1 are transcriptionally active in brain (H3K4me3 promoter peaks) and are within a neuronal highly methylated domain (N-HMD, defined as PMD in IMR90 but HMD in SH-SY5Y cells). In contrast, BDNF is inactive but poised for transcriptional activity differentially at different promoters and in cell lines and the alternative BDNF promoters are within a PMD in both SH-SY5Y and IMR90 cells. The nuclear lamina, a heterochromatic protein matrix at the nuclear periphery made of lamin and scaffold proteins, overlaps with both PMDs and light grey “off the map” locations in the chromatin state maps. CTCF (blue) shows multiple distinct sites genome-wide that are both intergenic and close to promoters and ubiquitous as well as tissue-specific.