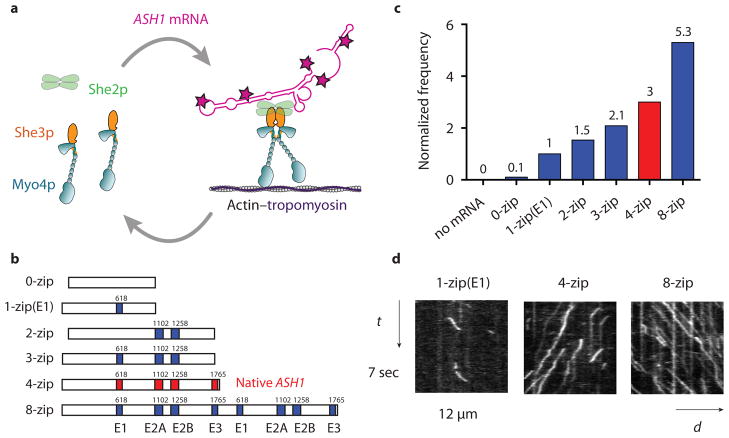
Figure 1.
The frequency of mRNP processive runs increases with zipcode number. (a) Diagram showing the She2p and mRNA dependent dimerization of Myo4p–She3p at 140 mM KCl. Motion of the motor-mRNA complex is visualized by incorporating Alexa Fluor 488-5-dUTP (indicated by red stars) into the ASH1 mRNA transcript. (b) Schematic of ASH1 mRNA constructs containing the indicated number of zipcodes. The four zipcode elements in native ASH1 mRNA (E1, E2A, E2B, E3) are indicated in red. The initial base position of each element in the native ASH1 sequence is indicated. The 0-zip construct has no zipcodes. The 1-zip(E1) construct contains only the E1 motif, 2-zip both the E2A and E2B zipcodes, and 3-zip the E1, E2A, and E2B zipcodes, all in their native positions. The 8-zip construct is formed from two concatenated 4-zip sequences. (c) Run frequencies of Myo4p mRNP motility from a representative experiment comparing ASH1 mRNA sequences containing varying numbers of zipcodes. The run frequency (number of runs per μM Myo4p per μm actin per sec) with 1-zip(E1) is normalized to one. The actin track has Tpm1p bound. Motion of the Myo4p motor complex without mRNA is visualized with She2p-YFP. The native ASH1 mRNA run frequency is indicated with a red bar. For ASH1 transcripts containing zipcodes, n ≥ 198, without zipcodes, n=13. (d) Kymographs of Myo4p mRNPs containing ASH1 mRNA with the indicated number of zipcodes. The slope of the trace is the speed of the mRNP. The length of the trace is its run length. The number of traces is related to run frequency. Conditions: 140 mM KCl, pH 7.4, 1 mM MgATP