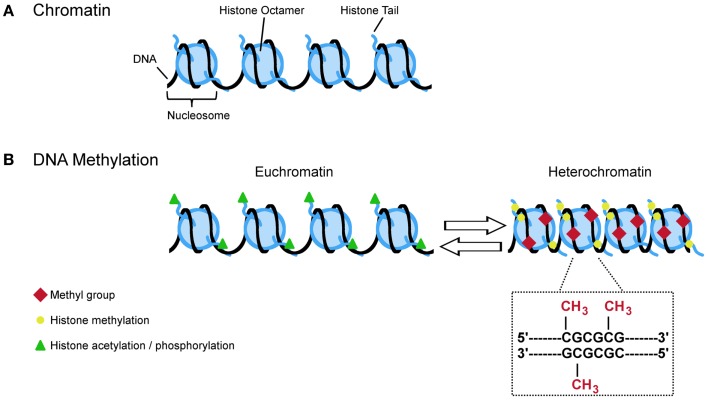
Figure 3.
A (simplified) guide to DNA methylation. (A) Approximate 147 bp of DNA are wrapped around the nucleosome which is made up by the core histones H2A, H2B, H3, and H4. The free ends of the histone tails serve as substrate for posttranslational modifications (e.g., acetylation, phosphorylation, and methylation) that influence the configuration of chromatin. Opened states (euchromatin) allow transcription while closed states (heterochromatin) restrict the transcriptional machinery. (B) Euchromatin is characterized by acetylation and phosphorylation (green triangles) whereas methylation (yellow circles) is more often found at heterochromatin. Specific histone modifications and DNA methylation reciprocally influence each other in deposition. For example, histone methylation at H3K9, H3K27, and H4K20 promotes DNA methylation at CpG dinucleotides. This covalent modification refers to the transfer of methyl group to cytosine resides at gene regulatory regions. Hypermethylation typically links to lasting transcriptional repression while hypomethylation favors gene expression.