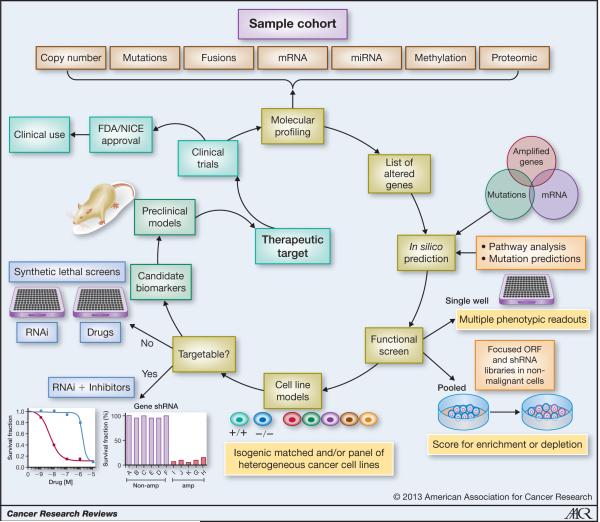
FIGURE 1. Schematic of data integration to identify therapeutic targets.
Molecular profiling of a cohort of primary breast cancers allows the identification of genomic alterations. Integration of this data is useful to identify potential oncogenic and tumor suppressive events in silico; e.g. genes that are overexpressed when amplified, in-frame fusion genes where the 3′ partner is overexpressed, mutations that are expressed at the mRNA level; genes that are under-expressed when deleted or methylated, disrupting fusions/structural rearrangements or mutations resulting in under-expression respectively. Additional in silico analyses can be performed to identify potential candidate driver genes by using prediction algorithms that ascribe biological meaning to genomic data. For example searching for significantly altered pathways that are more likely to contain driver genes, prediction of key transcriptional regulators of oncogenic programs and prediction of which missense mutations are likely to have a biological effect on the protein. Construction of cancer-focused screens can be a useful tool to investigate which candidate driver genes confer tumor specific dependencies. Oncogenic drivers and tumor suppressors are simultaneously assessed by constructing parallel libraries of cDNA ORF’s and shRNA’s, which are expressed in pre-malignant cells, and subsequently assayed for tumorigenicity either in vitro through the use of 3D models or in vivo. In addition, other measures of phenotypic alterations can also be assayed in tandem e.g. invasion, migration and resistance to anoikis. Both pooled screening using next generation sequencing for deconvolution and identification of biologically active ORF’s and shRNA’s, or single well screening can be used. To take forward hits from these screens, the use of appropriate cancer cell line models constitutes a more translationally relevant platform for drug discovery and development. Either a panel of phenotypically matched breast cancer cell lines (ER, PR, HER2, TP53) with and without the aberration of interest are used, or an isogenic cell pair to investigate the selectivity of the genomic alteration of interest. Genes that are directly targetable with validated inhibitors (e.g. kinases) can then be taken forward for further evaluation in the cell line panel e.g. for oncogenic events assessment with RNAi and available inhibitors, can be used to assess tumor dependency (cells with the aberration will be sensitive to gene inhibition, whereas those without will not). Aberrations that are not directly targetable with available inhibitors can be assessed through synthetic lethal screens using siRNA druggable libraries and drug screens. Candidate dependencies can be subsequently validated in pre-clinical models before evaluation in clinical trials. By identifying the genetic alteration and then identifying ways of targeting it, allows the genomic biomarker to be established a priori, cutting down the time to identify biomarkers of sensitivity during the drug development process. As is sometimes the case, promising preclinical data do not readily transfer to positive outcomes from early clinical trials. While inter- and intra-tumor genetic heterogeneity almost certainly play a role in resistance to targeted therapies, other molecular mechanisms can be teased out using the same approach described above. We can change the cohort of samples interrogated with molecular profiling to identify biomarkers for resistance or sensitivity to the targeted agent in question (i.e. before or after treatment, responders or non-responders). Identification of targets that are selective to inhibitors already in clinical trials will enhance the time to routine clinical use.