Figure 6.
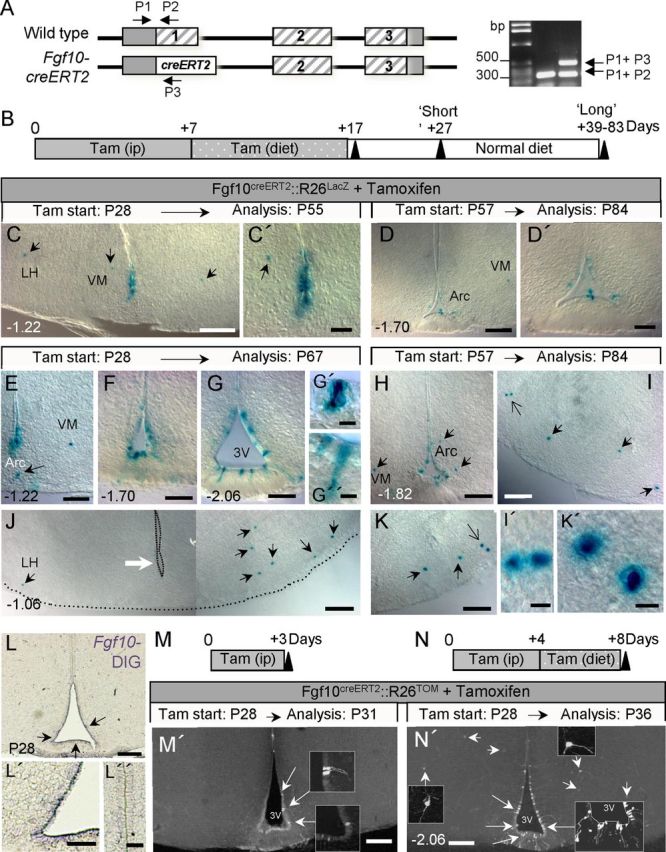
Direct lineage tracing of Fgf10+ tanycytes using Fgf10creERT2 and R26LacZ reporter mice. A, Schematic of Fgf10 gene locus showing its coding exons (hatched bars), the knock-in creERT2 transgene, as well as the position of genotyping primers (P1–P3) and the resulting PCR products, used to distinguish between the two alleles. B, Strategy for tamoxifen application and time course analysis of tamoxifen-treated animals. C–K', Images of recombined (Xgal+) cells in the hypothalamus of tamoxifen-treated Fgf10creERT2:R26LacZ mice analyzed at short (C–D') or long (E–K') time-points after the last tamoxifen dose. Large white arrow in J points to the absence of recombined tanycytes at bregma −1.06, contrasted with the presence of Xgal+ cells in the neighboring parenchyma (black arrows). Large open arrows in I and K point to pairs of Xgal+ parenchymal cells, magnified in I' and K', respectively. L–L”, In situ hybridization using DIG-labeled probes shows expression of Fgf10 by ventral ependymal cells (L, L', arrows) but neither by dorsal ependymal (L”) nor by neighboring parenchymal cells at P28. M–N′, Very short term recombination analysis after 3 (M, M') or 8 (N, N′) d of tamoxifen-treatment of Fgf10creERT2:R26TOM mice. Recombination is restricted to tanycytes after a 3 d treatment (large arrows and insets in M') while both recombined tanycytes (large arrows) and parenchymal cells (small arrows) are evident following an 8 d pulse (N′). Scale bars: C, D, H, L, M', N′, 100 μm; C', D', E–G, I–K, L', 50 μm; L”, 25 μm; I', K', 5 μm.
