Abstract
The ability to precisely deliver molecules into single cells is of great interest to biotechnology researchers for advancing applications in therapeutics, diagnostics, and drug delivery toward the promise of personalized medicine. The use of bulk electroporation techniques for cell transfection has increased significantly in the last decade, but the technique is nonspecific and requires high voltage, resulting in variable efficiency and low cell viability. We have developed a new tool for electroporation using nanofountain probe (NFP) technology, which can deliver molecules into cells in a manner that is highly efficient and gentler to cells than bulk electroporation or microinjection. Here we demonstrate NFP electroporation (NFP-E) of single HeLa cells within a population by transfecting them with fluorescently labeled dextran and imaging the cells to evaluate the transfection efficiency and cell viability. Our theoretical analysis of the mechanism of NFP-E reveals that application of the voltage creates a localized electric field between the NFP cantilever tip and the region of the cell membrane in contact with the tip. Therefore, NFP-E can deliver molecules to a target cell with minimal effect of the electric potential on the cell. Our experiments on HeLa cells confirm that NFP-E offers single cell selectivity, high transfection efficiency (>95%), qualitative dosage control, and very high viability (92%) of transfected cells.
Keywords: electroporation, electropermeabilization, transfection, single cell, microfluidic probe, nanofountain probe
Recent technological and scientific advances in cell biology techniques have enabled studies with single-cell resolution, and therefore, brought a paradigm shift in the fields of cell genomics and proteomics [1, 2]. Considering that heterogeneity is an intrinsic property of even seemingly similar populations of cells, studying cellular response to a particular gene or drug with single-cell resolution is a promising method to address such intrinsic heterogeneity [3, 4]. Consequently, in concert with progress in single-cell analysis techniques, methods to nondestructively target and inject/transfect single cells are also actively being developed [5].
In order to study the effect of a particular gene or drug on cellular behavior, the target molecule must be delivered into the cell without causing significant damage. For studies of spatial and temporal regulations within a cell, one of the most efficient methods is transfection with macromolecules such as DNA and RNA [6–10]. This allows monitoring the localization or biochemical state of proteins of interest, toward the goal of identifying mechanisms that control a living cell [11, 12]. Cell membranes, though only a few nanometers thick, effectively separate the cell cytosol from the extracellular buffer, and except for some small molecules, is largely impermeable to polar substances such as DNA, peptides, proteins, most ions, and large biomolecules. Current methods for delivery of molecules into cells include: microinjection using glass micropipettes [13–15] or nanostructures such as nanowires and nanotubes [16–19], nanostraws and stealth probes [20, 21], or fluidic probes [22]; carrier-mediated delivery using lipids, viruses, conjugated nanoparticles, or nanodiamonds [23]; and transient permeabilization of the cell membrane by electroporation [24]. Each method has its own advantages and disadvantages. For example, microinjection using glass micropipettes is applicable to most cell types and works for essentially any molecule size. However, successful and reproducible microinjection is highly user dependent and time consuming [25], rendering the method technically demanding and inefficient. Carrier-mediated delivery is limited to certain sizes and types of molecules, is often cell specific, and is only applicable for a population of cells, i.e., it is not practical for single-cell delivery. Furthermore, the carrier can cause undesired mutagenesis of the cell, which may skew measurements or be unsafe for human studies [26]. Electroporation enables delivery of biomolecules into a cell by creating transient and reversible nanopores in the cell membrane, increasing the cell membrane permeability and facilitating transmembrane transportation [27]. The nanopores are generated by application of an external electric potential to the cell membrane such that the transmembrane potential exceeds the dielectric breakdown-voltage of the lipid bilayer. Electroporation is applicable for both bulk [27, 28] and single-cell [26, 29] treatments, and is compatible with most cell types and sizes and types of agents. Thus, it is a flexible and powerful alternative to other intracellular delivery methods [26].
Bulk electroporation was first introduced in 1982 for delivery of DNA into mammalian cells [30]. In bulk (or batch) electroporation, a suspension of numerous cells, often on the order of 105–106, is loaded in a cuvette and placed in a device that applies a large electric potential (often in kV range) to the entire contents of the cuvette. The molecules to be transfected are also loaded into the cuvette and are taken up by the cells via diffusion upon generation of nanopores in the cell membrane. This bulk treatment results in different cells being exposed to the threshold breakdown-voltage at different times; therefore, control of the permeabilization and exact delivery dosage is not possible. In addition, the large required voltage is often toxic to cells due to excessive heat generation in the buffer [26].
Increasing interest in single cell studies has prompted the development of microscale electroporation and single cell electroporation (SCEP) techniques [26, 29]. Current SCEP techniques utilize microelectrodes [24, 31], micropipettes, electrolyte-filled capillaries [32–38], and microfabricated devices and microfluidics [39–53]. The first SCEP technique was implemented in 1998 by placing two carbon microelectrodes in proximity (within microns) to a single cell [24]. An electric potential was applied between the electrodes, and the agent in the media transferred into the cell through diffusion. The first microfabricated single-cell electroporation chip was reported in 2001 [39]. Using microfabricated chips, cells are either transferred through microfluidics to the vicinity of the electrodes or directly plated on the chip containing the electrodes. Because the electrodes are fixed in these chips, electroporation is limited to only the cells near the electrodes. A similar method is based on microfluidic electroporation in which the electric potential is applied through a conductive electrolyte to a cell trapped inside a microchannel [40]. Micropipette-based SCEP in intact tissues was reported in 2001 [32] using a micropipette filled with a conductive solution to electroporate a single cell positioned in contact with the tip of the micropipette. An atomic force microscope (AFM) has also been used recently for SCEP by placing a conductive AFM tip in contact with a target cell adhered to a conductive substrate. Biomolecules in the media transferred into the cell through the nanopores by diffusion [54].
Development of single cell electroporation instrumentation and protocols for practical use in biotechnology, drug discovery, and personalized therapeutics could transform the future of these fields. Despite significant progress in the development of SCEP techniques, a universal device that is easy to use, gentle to cells, precise, and scalable for higher throughput is lacking. Toward this goal, we report here a novel SCEP technique, called nanofountain probe electroporation (NFP-E), for the delivery of molecules into single cells. The nanofountain probe (NFP) is a microfabricated chip [55, 56] consisting of an array of cantilevers with an embedded microfluidic channel system and dispensing fountains. We first consider, theoretically and numerically, the electrical response of the NFP-E system resulting from the probe tip geometry and its position relative to the targeted cell membrane. We then experimentally demonstrate the capabilities of NFP-E by transfecting human cervical cancer (HeLa) cells with fluoraphore-labeled dextran and monitoring their viability using propidium iodide. We identified optimal NFP-E experimental parameters by monitoring the onset of contact between the NFP tip and a cell, using either optical observations or electrical measurements, and controlled transfection dosage by varying the duration of the applied input voltage.
NFP System for Single Cell Electroporation
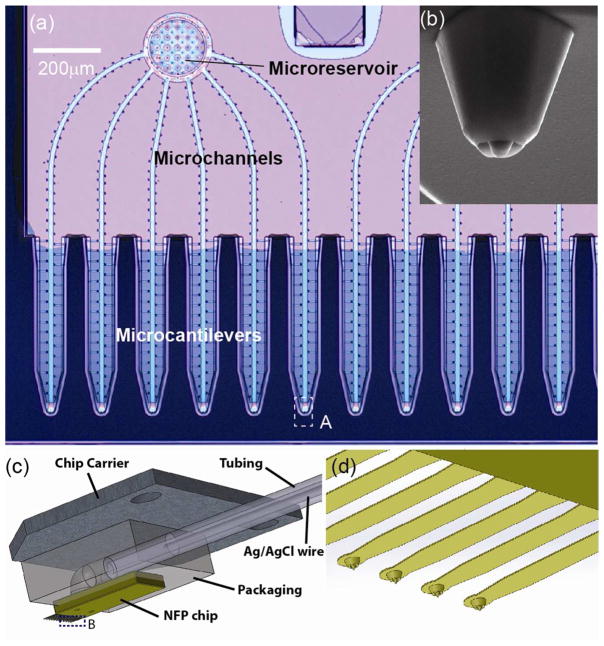
The geometric features of the NFP chips and the packaging used for the NFP-E system are shown in Figure 1. The NFP chip consists of twelve cantilevered probes connected to two independent micro-reservoirs through sealed microchannels (Figure 1a) [22, 55]. The NFP was previously used for parallel nanopatterning of several molecular inks and proteins [57, 58] and more recently for cell injection and cell substrate patterning to assess the effect of chemotherapy drug dose on cell death [22]. In the NFP, the molecular solution to be delivered or patterned is stored in the on-chip microreservoirs and loaded into the microchannels using capillarity or an externally applied pressure. Due to the small probe aperture (about 750 nm) and its precise position control, by means of a micromanipulator or an AFM, biomolecules can be transfected to a specific region of interest on a target cell (as a reference, typical in plane cell size is on the order of 10–20 μm). Further details on microfabrication, analysis, and application of the NFP can be found in [22, 55, 56].
Figure 1.
NFP chip and packaging for single cell electroporation: (a) optical microscopy image of the NFP with multiple probes for parallelized single cell electroporation; (b) SEM image showing a zoom-in view of the probe tip (region A in (a)); (c) schematic of the packaged NFP chip including the chip carrier, polycarbonate packaging, plastic tubing and silver/silver chloride wire; (d) magnified view of the cantilevers in region B of (c).
For easy assembly and handling, the NFP chip is packaged onto a polycarbonate block with a built-in channel that is affixed, on the other side, to a metallic chip carrier (Figure 1c and S1b in Supporting Information). The metallic chip carrier can be easily mounted on either a nanomanipulator or an AFM for position and force control. To apply external electric potential and pressure, the packaging is connected to a plastic tube containing an internal Ag/AgCl wire and filled with a conductive solution. One opening of the tube is connected to an external microfluidic pump (Femtojet, Eppendorf, Germany), while the other opening is sealed with the inserted Ag/AgCl wire as shown in Figure 2a. This integrated packaging allows delivery of the loaded solution to the NFP tips with precise flow rate control and provides a method to apply the desired electric potential to a cell for single cell electroporation.
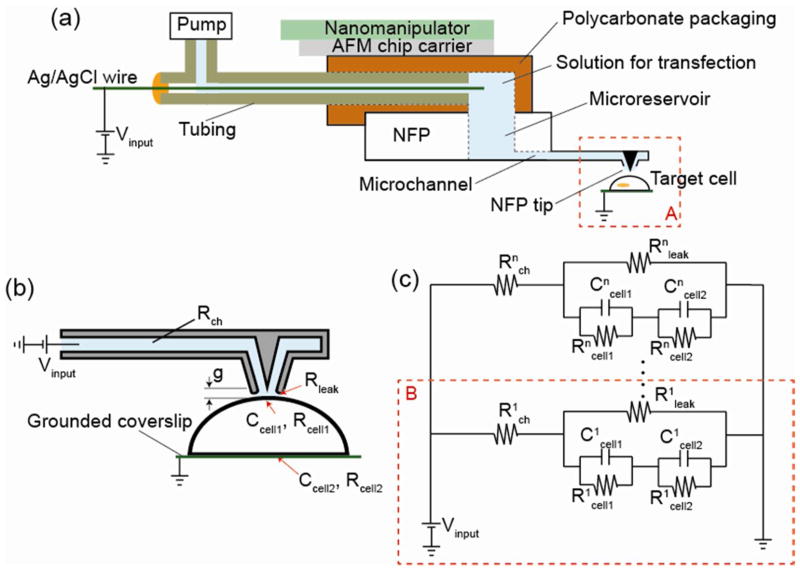
Figure 2.
The electrical circuit of the NFP-E system during electroporation: (a) schematic of the packaged NFP for single cell electroporation; (b) schematic of the equivalent electrical circuit of the NFP-E when an NFP tip is in contact with a cell; (c) lumped model representation of an n-array circuit for an n-probe NFP chip, which is equivalent to a parallel model of a one-probe circuit.
Although the NFP can be used with a standard AFM system as previously demonstrated [22, 56], here we use a nanomanipulator (InjectMan NI 2, Eppendorf, Germany) to control three dimensional displacement of the NFP probes with step size resolution of 40 nm (see Figure S1a in Supporting Information). The nanomanipulator is mounted on an inverted fluorescence microscope (Nikon Eclipse Ti-U) equipped with a color CCD camera (Jenoptik CFcool, Germany).
For real time monitoring of live cells, during electroporation, an inverted fluorescence microscope is employed. Adherent cells are cultured on a coverslip coated with a conductive thin film, e.g., Cr/Au, and placed in a liquid cell (Park Systems, CA) on the microscope sample stage. Once a target cell is optically selected, the NFP probe is displaced, using a nanomanipulator, such that the NFP tip covers the cell in a region of interest. The onset of contact is detected either by optical observations of the cell morphology or by the change in electrical resistance due to the sealing between probe tip and cell. After contact, an electric pulse is applied between the Ag/AgCl wire and the conductive coverslip to induce electroporation. The external input signal is generated by a pulse generator (Standard Research Systems DS345), amplified by a voltage amplifier (OPA445, Texas Instruments), and monitored using an oscilloscope (LeCroy 9384L). The electrical resistance of the circuit during electroporation is monitored using a digital multimeter (Agilent 34401A). Further details on the experimental procedures are given in a subsequent section.
Analytical and Numerical Model for NFP-E
Electroporation is a three-stage process: membrane charging, pore nucleation, and pore evolution [59]. The charging step takes several microseconds [60] until the transmembrane potential reaches a certain threshold (often 0.2~1 V [26]), which triggers the onset of nanopore nucleation on the cell membrane. After pore nucleation, some of the pores grow to a radius of approximately 15 to 25 nm; however, the majority of the pores remain with a size of several nm in radius. The time for pore size evolution is on the order of milliseconds and is driven by energy minimization of the whole cell membrane [59]. After the electric pulse is turned off, the cell membrane discharges through the existing pores and the transmembrane potential returns to zero. Consequently, the pores shrink to the minimum energy state (0.8–1 nm), and finally reseal on the order of seconds [26, 59]. Based on theoretical models, the number of pores (generally small pores) increases with the strength of the applied pulse [26, 59]. For example, a pulse of 1.4 V with duration of 10 ms creates about 3×105 pores with an average radius of ~9 nm. According to theoretical models based on energy principles, pore density can be as high as 109/cm2, with 97% of the pores having a radius of about 1 nm [60].
To trigger formation of nanopores on the cell membrane, needed for successful electroporation, a transmembrane electric potential in the range of 0.2~1 V is required. Hence, we theoretically investigate the electric potential drop across a cell membrane when using the NFP geometry. We start by modeling a single NFP tip in contact with a single cell membrane, as shown schematically in Figure 2a and 2b. In the lumped model, the input voltage is Vinput, while Rch and Rleak represent the resistance, along the microchannel between the Ag/AgCl wire and NFP tip, and between the cell membrane and NFP tip, respectively. The leak resistance, Rleak, is a function of the probe-cell membrane gap, g, given that such resistance is inversely proportional to the leakage cross-sectional area. The cell is modeled as a half-sphere with a 10 μm radius. When the cell is subjected to an electric field, the cell membrane behaves as a capacitor and resistor in parallel [61]. The cell membrane is divided into two parts: the part in contact with the NFP tip (Rcell1, Ccell1) and the part of the cell adhered to the grounded substrate (Rcell2, Ccell2). Note that Rcell1 is much larger than Rcell2 because the surface area of the cell membrane in contact with the NFP (Rcell1) is much smaller (1%) than the area of contact with the substrate electrode (Rcell2). The cell cytosol has much lower resistance compared to the lipid bilayer, and therefore, its resistance is neglected. The electrical conductance of the liquid in the NFP microchannels is defined as G = σ A/l, where σ is the electrical conductivity of the liquid, and A and l are the cross-sectional area and length of the microchannel, respectively.
The microchannel is connected in series to the cell and surrounding media through the gap g while the cell membrane and g are connected in parallel. Therefore, the one-probe NFP in Figure 2b is represented by an equivalent lumped model shown in region B of Figure 2c. Note that the resistance of the surrounding conductive media between Rleak and the electrical ground at the coverslip is ignored since the surrounding media has a much smaller electric resistance than other electrical components in the system due to having larger cross section. Each probe on the multi-probe NFP shown in Figure 1a is connected to a reservoir in parallel and hence an NFP with n-probes can be represented by an n-array of one-probe circuits in parallel (Figure 2). For simplicity, in the analysis here discussed, we only consider the steady-state response for a constant input voltage, Vinput, such that capacitive effects, e.g., at the cell membrane, can be ignored. Note that the potential drop through each probe on an n-probe NFP is Vinput = V1 =…= Vn where Vi is the potential drop through the ith probe (i=1, 2, …, and n). The same potential drop through each probe indicates that parallelized electroporation can be implemented without applying higher voltage than the one it would be required for single-probe electroporation.
Next we explore the relationship between the transmembrane potential drop, Vm, and system parameters such as Rch, Rleak, and Vinput. Such analysis is important to determine optimal electroporation conditions and more specifically, requirements for probe-membrane interactions. As previously discussed, the transmembrane potential drop from each individual probe is independent of other probes; therefore, this analysis is presented in the context of a single probe NFP system as depicted in Figure 2c, region B. Note that for a given Vinput, there is a potential drop through the microchannel (Vch), due to Rch, and a further potential drop through the cell (Vcell). From the single probe circuit, region B in Figure 2c, it can be shown that the normalized potential drop through the microchannel (Vch/Vinput) can be written as
| (1) |
where Rcell = Rcell1 + Rcell2. Note that Rcell/Rch ≫ 1 due to the small conductivity coefficient (5.0×10−7 S/m [61]) of the cell membrane and the small contact area between the NFP tip and cell membrane. Equation 1 shows that Vch/Vinput → 0 for Rleak/Rch ≫ 1 while Vch/Vinput → 1 for Rleak/Rch ≪ 1. This result indicates that the potential drop through the cell membrane increases with larger Rleak or equivalently with smaller probe-cell membrane gap g.
Even when Eq. (1) provides qualitative understanding of NFP-E, the model does not predict quantitative effects on the local electric potential field near a target cell. An accurate prediction of such local field is essential to determine the critical applied voltage for nanopore formation and probe location with respect to the cell membrane. Unlike bulk electroporation where each individual cell is exposed to a uniform electric field, the electric field for single cell electroporation is often non-uniform and hence an analytical solution is often not feasible [60]. Therefore, we used COMSOL Multiphysics to simulate the distribution of the electric potential through a cell membrane. The results are summarized in Figure 3. The numerical model consists of the NFP positioned on top of a cell embedded in conductive media. For these numerical simulations, the partial differential equation ∇(σ∇V)=0 is used with appropriate boundary conditions, where V is the input electric potential and σ is the electrical conductivity [60]. The electric potential was applied to the liquid in the NFP fountain and the surface under the cell is electrically grounded. The cell membrane was modeled as a 5 nm layer enclosing a conductive media (cell cytosol). The NFP geometry was implemented as an insulating boundary condition. The parameters used in the analysis are given in Table 1. Moreover, the analysis used Vinput = 10 V. With this model, we quantitatively evaluated conditions for the applicability of the conclusions drawn using the lumped electrical models, i.e., (i) that the potential drop, on each probe, is independent from other probes, and (ii) that the transmembrane potential drop can be maximized by increasing Rleak.
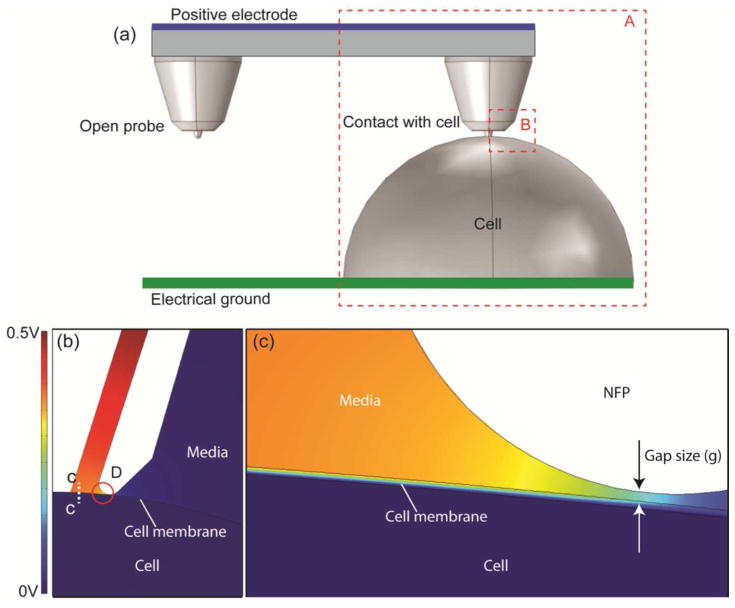
Figure 3.
Computational simulation of the electric potential distribution during NFP-E for a 10 V electric potential difference between the positive electrode and electrical ground; (a) three dimensional model of one-probe (region A) and two-probe NFP systems; (b) electric potential field of the cross section of region B in (a); (c) a magnified view of region D in (b).
Table 1.
Parameters used for numerical analysis of the NFP-E [61]
| Definition | Value |
|---|---|
| Membrane conductivity | 5×10−7 S/m |
| Intracellular conductivity | 0.45 S/m |
| Extracellular conductivity (the buffer) | 1 S/m |
To examine potential drop independence from probe number, we compared two different cases: (1) a single-probe NFP with g = 8 nm (region A in Figure 3a) and (2) a two-probe NFP with one open probe and one with g = 8 nm (Figure 3a). For the two-probe NFP case, both probes are electrically connected to a microreservoir like in the actual NFP chip. Figure 3b shows the potential field at the cross section of region B in Figure 3a for each case, and Figure 3c shows a magnified view of the region denoted as D, in Figure 3b. The maximum transmembrane potential drop is observed to occur in the area directly covered by the NFP, and the electric field is localized to the region in close proximity to the probe tip. To compare the transmembrane potential drop for the one- and two-probe cases, the potential drop profiles along C…C in Figure 3b are shown in Figure 4a. Examination of the results reveal that the potential drop through a cell membrane, for the probe in contact with the cell, is 0.38 V in both cases, confirming that electric potential drop through each probe is independent of the other probes. It is worth noting that, although demonstration of parallelized cell electroporation is not the main focus of the present manuscript, this is an important conclusion for parallelized single cell electroporation because it would be impossible to achieve identical cell-probe contact for each probe on a parallel NFP chip due to slight variations in the NFP probe geometries and heterogeneity of cell size and shape. In addition, as mentioned earlier, the parallelized electroporation does not require higher voltage than single-probe electroporation which is also an important feature for parallelized electroporation because the magnitude of the applied voltage directly correlates to cell viability [26].
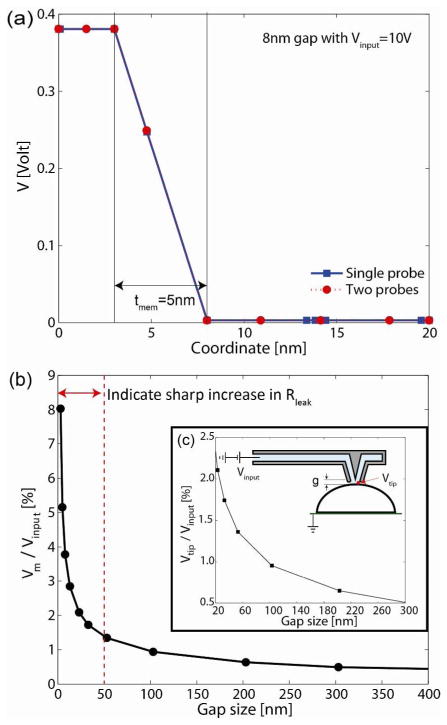
Figure 4.
Numerical simulations of the transmembrane potential drop through a cell membrane (C…C in Figure 3b). In (a), the gap between the NFP and cell membrane is 8 nm for both one- and two-probe NFP systems, and the transmembrane potential drop for both cases is identical despite the fact that the two-probe NFP case has one opened probe (as shown in Figure 3a). This confirms independence of potential drop in each probe. In (b), the transmembrane potential drop, normalized by the input voltage, Vinput, is plotted as a function of gap size (g). (c) Local voltage at the tip (Vtip) as a function of g for a given input voltage, Vinput.
To study the effect of gap size (g) on transmembrane potential drop during electroporation, we again modeled the one-probe NFP system since we concluded that potential drop through each probe is independent. The normalized transmembrane potential, V*m = Vm/Vinput, through C…C in Figure 3b is shown as a function of g in Figure 4b. We present the results in normalized form. Note that because the governing equation is linear, the simulation results can be applied to any electric potential input. As expected, the normalized transmembrane potential V*m sharply increases as g decreases, whereas it approaches zero asymptotically as g increases with the major potential drop then occurring across the chip microchannel. For example, in order to trigger the onset of formation of nanopores (Vcr = 0.2~1 V), the needed far field input voltage is Vinput = 3.0~19.4 V at g = 5 nm (V m= 5.2%) while Vinput = 9.6~48 V at g = 23 nm (V*m = 2.1%). This shows that lower input voltage can be used for NFP-E by decreasing g. It is worth mentioning that achieving an extremely small g could subject a cell to a force that might stress or damage it. This could be the case because any nanoscale local asperity or irregularity in the shape of the NFP tip may introduce a substantial change in the Rleak. Note that a sharp increase in V*m is predicted when g<50 nm, Figure 4b. Hence, the prediction suggests that when the NPF approaches a target cell, the rate of increase in Rleak, at small g, could be used as a detection scheme for detecting the onset of NFP-cell contact. We will discuss below experimental results that demonstrate the feasibility of the proposed method.
It is important to note that local transmembrane voltage (Vm) is much smaller than the input voltage (Vinput), see Figure 4b and 4c. Therefore, even when a seemingly high input voltage is applied to NFP system, a target cell is subjected to only a small fraction of the input voltage. For example, a Vm = 0.63V is achieved when Vinput = 30 V and the gap size g = 23 nm. Consequently, a minimal effect due to the electric potential is expected during NFP-E. In addition, the size of the NFP tip is much smaller than the area of the cell in contact with the substrate; therefore, the major transmembrane potential drop occurs only in the area in close proximity with the NFP tip. Hence, nanopores on the cell membrane will form only in a localized region while the rest of the cell membrane will remain intact. The well-controlled local voltage at the NPF tip (Vtip) and highly focused electric field are unique features of NFP-E compared to bulk electroporation. These result in a comparatively much higher transfection efficiency and cell viability. Note that it is difficult to directly control the gap size within 30–50 nm because the length scale of local asperity and irregularity in the shape of the probe become relevant within such a small gap. Therefore, the focus of the gap size study was to identify a practical method to detect the onset of probe-cell contact to ensure gentle mechanical contact.
SCEP Experimental Results
Models of the NFP-E system indicate that (1) the electric potential drop through each probe is independent, (2) parallelized electroporation with multiple probes does not require higher input voltage than single probe electroporation, (3) the transmembrane electric potential drop increases with larger input voltage and smaller gap between the NFP tip and cell membrane, (4) the NFP creates a highly focused electric field only within a small region of interest, and (5) local voltage at the tip is much smaller than the input voltage. To validate such predictions, we performed single cell electroporation experiments on HeLa cells using the NFP-E system. We obtained HeLa cells from the American Type Culture Collection (ATCC #CCL-2) and cultured them in Dulbecco’s Modified Eagle Medium (SIGMA) with L-glutamine and phenol red as pH indicator, supplemented with 10% FBS (SIGMA) and 1X penicillin/streptomycin (SIGMA). The cultured cells were maintained in a humidified incubator at 37 °C and 5% CO2. For electroporation experiments, the cells were plated the day before the experiment on a round 25 mm glass coverslip with a thin Cr/Au film and incubated in DMEM media. The thin metal film acts as one of the electrodes in electroporation experiments. The thickness of the coating was chosen to ensure both low resistance and good transparency for imaging cells using an inverted optical microscope. On the day of the experiment, the coverslip with plated cells was rinsed multiple times with DMEM without phenol red to avoid autofluorescence during fluorescence imaging of the cells. The coverslip was then placed in a liquid cell (Park Systems) and imaged using the inverted optical microscope, while DMEM media without phenol red was added to maintain the cells submerged throughout the electroporation experiments.
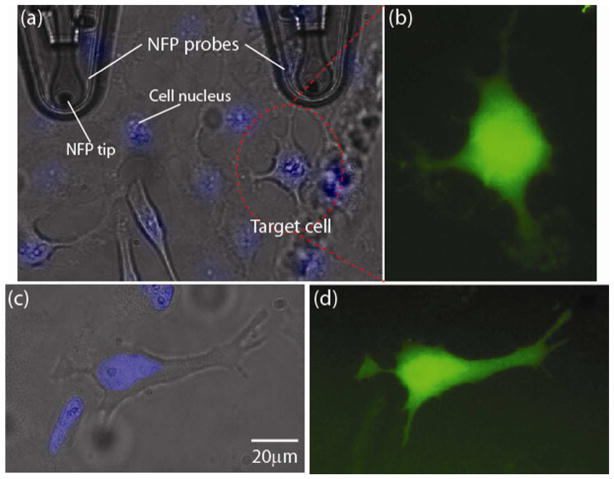
In order to monitor the electroporation efficiency, we transfected cells with 3000 MW fluorophore-labeled dextran (Alexa Fluor 488, Life Technologies). The labeled dextran was diluted with water to a working dilution of 1 mg/ml. Once the dextran solution was added to the packaged NFP chip through the tubing (Figure 1c), external pressure was applied until the dextran solution filled the microreservoir. Microchannels connecting the reservoir to the probe tips were quickly filled by capillary forces (see Figure S1c in Supporting Information). At this point, the pump was turned off. The probe filling was confirmed by optical observation and/or by electrical measurement between the two electrodes as depicted in Figure 2a. For example, we measured a sudden change in resistance (from open circuit to 10~20 MΩ for the multi-probe NFP) when the conductive media reached the tips. While imaging HeLa cells, using regular 10X and phase 40X objectives to identify a target cell (Figure 5a), a probe tip was positioned in contact with the target cell followed by single cell electroporation and delivery of dextran molecules into the cell. Based on our parametric study, summarized in Table 2, during the NFP-E experiments we used input voltages in the range of 15–30 V and square wave signals at 200 Hz. Examples of single cell electroporation using the described protocols are given in Figure 5, which shows successful delivery of Alexa Fluor 488 into the target cells.
Figure 5.
Transfection of dextran Alexa Fluor into a targeted HeLa cell by NFP-E at 30 V with 1 second input signal duration: (a) optical image of NFP tip and HeLa cells; (b) fluorescence image of the target HeLa cell in (a) after electroporation and transfection with dextran Alexa Fluor 488; (c) optical image of a second target cell; (d) fluorescence image taken 1 hour after electroporation of the target cell in (c). Note that the cell nuclei in (a) and (c) are stained by Hoechst 33342 (Invitrogen) for better single cell selection.
Table 2.
Parametric study of input voltage during electroporation.
| 0–7 V | 15–30 V | 40–60 V | |
|---|---|---|---|
| Electroporation | No transfection | Transfection | Transfection |
| Cell | No change | No change | Damaged |
| Image | Figure S2a | Figure 5 | Figure S2b and S2c |
The analysis previously discussed reveals that resistance change can be used to identify probe-cell contact. In this context, it is worth noting that when contact was assessed visually, there were instances in which a marked decrease in fluorescence intensity was observed after one hour, likely due to cell membrane damage (see Figure S2 in Supporting Information). Such damage was correlated to excessive contact force or input voltage. In these cases, the fluorescently labeled dextran began to diffuse out of the cells. In contrast, the cells shown in Figure 5 were imaged 1 hour after electroporation and they exhibited an undetectable change in the level of fluorescence intensity, implying that full recovery of the induced nanopores occurred.
As predicted by our modeling, one of the most important factors affecting the success rate of single cell electroporation experiments is gap control between NFP tip and cell membrane during the pulse application. If the probe tip is too far from the cell, the electric field would not be strong enough to produce membrane pores. On the other hand, if the probe tip is pushed too hard against the cell, it can damage it and eventually rupture the cell. To avoid these two scenarios, two different methods could be used. The first method is to optically monitor the cell morphology using the inverted microscope during the approach of the probe to the cell. This method is simple, but requires an experienced operator and it is difficult to automate. Alternatively, using the NFP, one can monitor resistance, in view that our model predicts a sharp increase in the circuit resistance as the NFP comes in close proximity to the cell membrane, i.e., g<50nm. In order to validate this prediction and to demonstrate a detection method based on an electrical measurement, easy to automate with appropriate software, we employed a single probe to experimentally measure the change in electrical resistance as the probe approached, contacted, and retracted from the cell membrane at a rate of 100 nm/s (Figure 6). The results confirm that a sudden increase in the electrical resistance of the circuit can be measured when the probe makes contact with the cell membrane. This sudden increase, up to 143%, is due to an abrupt change in the leakage resistance (Rleak) (Figure 2b) resulting from the opening of the probe tip being sealed by the cell membrane. The opposite effect was observed upon probe retraction. These experimental results clearly suggest that automated position control of the NFP probe, with respect to a target cell, can be implemented by resistance measurement in the NFP-chip circuit.
Figure 6.
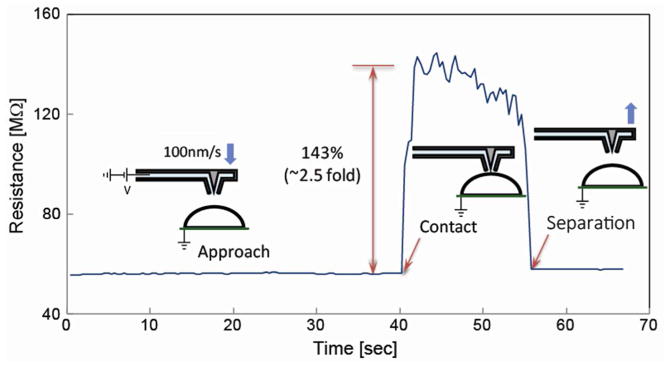
Resistance measurements as a function of NFP tip-cell membrane gap. The sudden increase in the electrical resistance of the NFP-chip circuit, upon contact with a cell, is observed. This feature can be exploited for automated detection of the NFP probe-cell membrane interaction.
As mentioned earlier, potential drop through each probe on a NPF chip is independent of the other probes; therefore, each probe can be used interchangeably during single cell electroporation. We experimentally confirmed the theoretical prediction. For example, we observed that a NFP probe was clogged after continuous use due to repeated interaction between probe and cells. Even when a particular probe was clogged, we could continue electroporation by switching to another probe on the same NPF chip without modifying any of the electrical input signals. The multiple parallel probes are a unique advantage of the NFP-E system in comparison to other microscale electroporation methods, e.g., micropipette based electroporation.
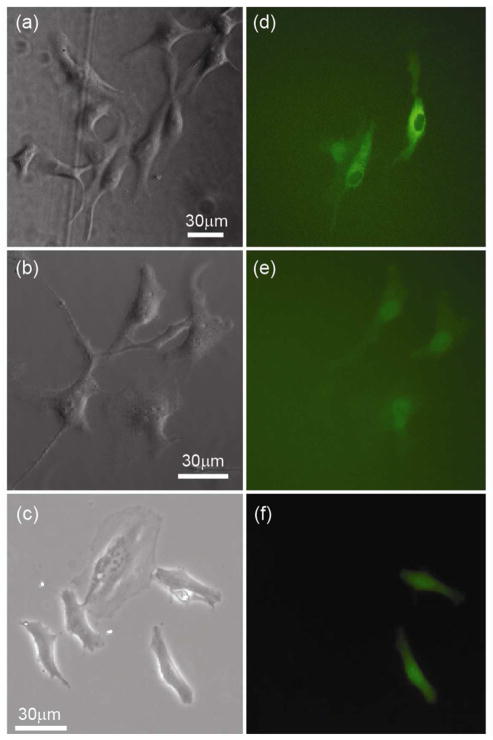
To examine the ability of the NFP-E to control dosage, the effect of input pulse duration was explored. From our parametric study of input voltage (Table 2), we concluded that the optimal range is 15–30 V when using a square wave at 200 Hz. Hence, we used a 30 V input signal and varied the pulse duration as the dosage control parameter (Figure 7). Cells labeled A and B, in Figure 7a, were first electroporated with two 0.25 sec input square wave signals with a 1 sec time interval between square waves. Cells C and D were then electroporated using two 0.5 sec square waves and 1 sec time interval between square waves. Finally, Cells E and F were electroporated with two 1.5 sec square waves with the same 1 sec time interval. Note that cells C and D (Figure 7c), electroporated with 1 sec duration, exhibit much brighter fluorescence intensity than cells A and B, electroporated with 0.5 sec duration. The same trend in the fluorescence intensity is observed in Figure 7d between cells C/D and cells E/F due to the increase in applied input pulse duration. The fluorescence intensities among cells exposed to the same signal duration show comparable and consistent intensity, indicating that dosage control can be achieved by varying control parameters during electroporation. Exact quantification of dosage is beyond the scope of this manuscript and it is left to future studies. Note that Figures 7a and 7b-d show bright-field and fluorescence images, respectively, with a consistent field of view, but the intensity range of the fluorescence images was rescaled to optimize contrast among electroporated cells.
Figure 7.
Single cell transfection of dextran Alexa Fluor 488 into targeted HeLa cells using the NFP-E at 30 V (square waves at 200Hz with 1 sec interval between pulses): (a) bright field image of the HeLa cells before electroporation; (b) fluorescence image after electroporation of cells A and B with two 0.25 sec signals; (c) fluorescence image after electroporation of cells C and D with two 0.5 sec signals; (d) fluorescence image after electroporation of cells E and F with two 1.5 sec signals.
To further evaluate the success of the single cell electroporation experiment, we have performed viability tests using propidium iodide (PI, eBioscience), one of the most commonly used protocols for viability assay. The results of this viability test are shown in Figure 8 where a target cell (cell A) was electroporated and transfected. The fluorescence image shown in Figure 8b indicates that cell A was successfully transfected with the fluorophore. After electroporation, the cells were kept in an incubator in complete DMEM media for 4 hours. After incubation, PI was used to stain cells to discriminate dead cells from live cells. Note that PI exhibits fluorescence in the red spectrum when it is bound to nucleic acids and that PI is only permeable in dead cells. Figure 8e reveals absence of red fluorescence, which indicates all cells in the field of view were alive. As shown in Figure 8d, the undiminished intensity of the green fluorescence from cell A indicates that the cell membrane fully recovered without diffusion of the transfected Alexa Fluor out of the cell. Our control experiment on dead cells (Figure S3 in Supporting Information) confirmed that the PI protocol for viability testing worked as expected. We repeatedly carried out viability tests (Figure S4 in Supporting Information) and proved that NFP-E is a superior and effective method with electroporation success rate in excess of 95% (Figure S5 in Supporting Information) and viability higher than 92%.
Figure 8.
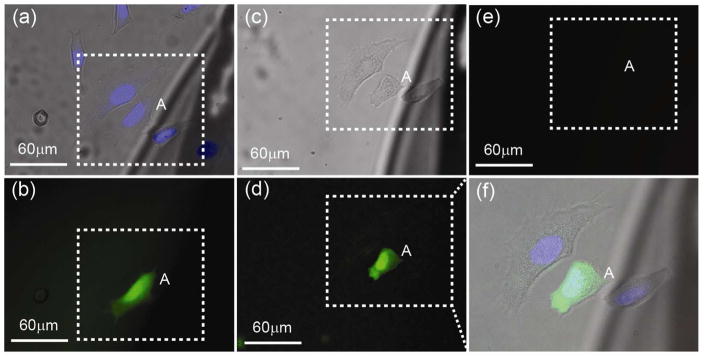
Live/dead assay using propidium iodide (PI). (a) Alexa Fluor was electroporated into target cell A. (b) Shows the corresponding fluorescence image. After electroporation, the coverslip was kept in an incubator for 4 hours and then stained with PI to image dead cells. (c)–(e) are bright field, green (Alexa Fluor), and red (PI) florescence, respectively, after PI staining. No red fluorescence was observed in (e), which indicates all cells in the field of view were alive. Note that (f) is the merged bright field and fluorescence zoom-in images in the region of interest (dotted box) after 4 hour incubation.
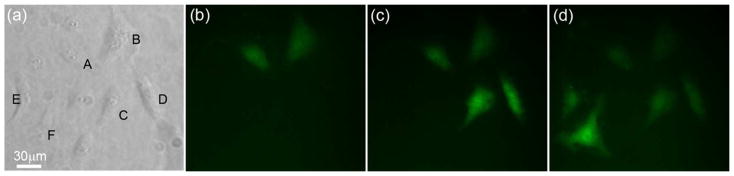
To further demonstrate the applicability of the NFP-E, we transfected HeLa cells with other biomolecules including (i) 70 kDa bovine serum albumin (BSA), (ii) 20 kDa GAPDH-target beacon, and (iii) 2 MDa plasmid DNA as shown in Figure 9. Bright field and corresponding fluorescence images of the cells are shown in Figures 9a–c and 9d–f, respectively. During NFP-E of BSA tagged with Alexa fluor 488, we observed that BSA could be delivered to the cytoplasm or directly into the nucleus (Figure 9d) by positioning the probe away from or on top of the nucleus, respectively. This capability is owed to the small size of the NFP probe tip, its precise position control, and localized nature of the applied voltage. Although nucleofection of single cells is beyond the main scope of the current manuscript, this result implies that the NFP-E can be used for biological studies where proteins need to be transfected directly into the nucleus. In addition to BSA, we transfected cells with small DNA molecules, in a hairpin configuration (GAPDH-target beacon), and much larger DNA molecules (GFP-expressing plasmids). Experimental results are shown in Figures 9b and 9c, respectively. The corresponding fluorescence images in Figures 9e and 9f indicate successful delivery of DNA into single cells. These studies show that the NFP-E technique is capable of transfecting molecules with various sizes and charges into single cells while maintaining their viability.
Figure 9.
Transfection of HeLa cells with protein and DNA via NFP-E. Bright field images are shown in (a)–(c) with corresponding fluorescence images to their right in (d)–(f). Transfection was achieved with (d) 70 kDa bovine serum albumin, (e) 20 kDa GAPDH-target beacon (30 base pairs), and (f) 2 MDa GFP-expressing plasmid DNA (~5000 base pairs). Note in (d) that delivery into the cytosol or directly into the nucleus was possible.
Conclusions
A robust and nondestructive method for controlled, in situ delivery of molecules into cells is needed to advance the state-of-the-art in personalized medicine and therapeutics. Development of SCEP instrumentation like the NFP-E system, and protocols for practical use in biotechnology research, drug discovery, and personalized therapeutics, could transform the future of these fields. Hence, demand is great for the development of an universal tool for single cell electroporation that is robust, easy to use, efficient, and gentle to cells.
Bulk electroporation is increasingly being used as the transfection method of choice despite being extremely disruptive to cells due to large heat generation from the kV-range applied voltage. In addition to toxicity, the bulk electroporation technique also suffers from issues such as lack of dosage control because the uptake of biomolecules after pore generation is governed by random diffusion, resulting in a heterogeneously transfected cell population. Thus, the technique is not suitable for applications involving sensitive cells (e.g., primary cells) that require high yield with precise cellular delivery (dosage). In contrast, we have demonstrated that the NFP-E system is minimally disruptive to cells, only a very small portion of the cell membrane is subjected to the electric field and probe-membrane contact can be detected electronically, so effective transfection was accomplished with applied input voltages of only ~30 V leading to transmembrane voltages Vm~0.6 V. Moreover, we demonstrated the ability to precisely control and monitor the contact force applied by the probe on the cell, using optical imaging and electrical detection, reduces stress and potential cell damage upon contact.
Another advantage of NFP-E is its compatibility with common microscopy methods such as AFM [22] as well as epifluorescence and confocal microscopy, which allows the entire transfection process and post-transfection cellular response to be monitored in an optical microscope. Further advantages include the small volume of each NFP microchannel (~3 pL) and the precise control of the delivery of these often expensive biomolecules or other agents. Because the molecules are confined in the NFP, the generated nanopores are exposed to a high concentration of transfection agent, minimizing the amount of consumable biomolecules compared to other methods. In addition, the NFP chips are fabricated at the wafer level so batch processing can be readily scaled up for mass production.
We have demonstrated here electroporation of single cells using NFP technology for the delivery of membrane-impermeable biomolecules into HeLa cells. The NFP-E system has unprecedented capabilities for targeted transfection such as single cell selectivity, high transfection efficiency, dosage control, and ultrahigh cell viability. The NFP-E tool has potential to enable novel biological studies including: (1) single cell analysis (gene expression studies, time-dependent cell biology, protein interaction studies, drug toxicity and response), (2) cell line development, and (3) stem cell reprogramming/differentiation.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation under award IIP-1142562 and the National Institutes of Health under award 1R41GM101833-01. We thank Professor Punit Kohli of the Department of Chemistry at Southern Illinois University for valuable discussions, Professor Dean Ho of the Department of Biomedical Engineering at Northwestern University for providing HeLa cells, Professor Gang Bao of the Department of Biomedical Engineering at the Georgia Institute of Technology for GAPDH-target beacon, and Rodrigo Bernal for assistance with the experimental setup.
References
- 1.Leslie M. The power of one. Science. 2011;331:24–26. doi: 10.1126/science.331.6013.24-a. [DOI] [PubMed] [Google Scholar]
- 2.Kalisky T, Quake SR. Single-cell genomics. Nature Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 3.Altschuler SJ, We LF. Cellular heterogeneity: Do differences make a difference? Cell. 2010;141:559–563. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nature Reviews: Molecular Cell Biology. 2011;12:119–125. doi: 10.1038/nrm3044. [DOI] [PubMed] [Google Scholar]
- 5.Andersson H, Berg Avd. Microtechnologies and nanotechnologies for single-cell analysis. Current Opinion in Biotechnology. 2004;15:44–49. doi: 10.1016/j.copbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Stephens DJ, Pepperkok R. The many ways to cross the plasma membrane. Proceeding of the National Academy of Sciences. 2001;98:4295–4298. doi: 10.1073/pnas.081065198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S, Nakamura C, Obataya I, Nakamura N, Miyake J. Gene expression using an ultrathin needle enabling accurate displacement and low invasiveness. Biochemical and Biophysical Research Communications. 2005;332:633–639. doi: 10.1016/j.bbrc.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Yu LC. Single-cell microinjection technology in cell biology. BioEssays. 2008;30:606–610. doi: 10.1002/bies.20759. [DOI] [PubMed] [Google Scholar]
- 9.Han S, Nakamura C, Kotobuki N, Obataya I, Ohgushi H, Nagamune T, Miyake J. High-efficiency DNA injection into a single human mesenchymal stem cell using a nanoneedle and atomic force microscopy. Nanomedicine. 2008;4(3):215–225. doi: 10.1016/j.nano.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Heiser WC, editor. Methods in Molecular Biology. Vol. 1. Humana Press Inc; Totowa, NJ: 2004. Gene Delivery to Mammalian Cells: Nonviral Gene Transfer Techniques. [Google Scholar]
- 11.Pinaud F, Clarke S, Sittner A, Dahan M. Probing cellular events, one quantum dot at a time. Nature Methods. 2010;7:275–285. doi: 10.1038/nmeth.1444. [DOI] [PubMed] [Google Scholar]
- 12.Ashley CE, Carnes EC, Phillips GK, Padilla D, Durfee PN, Brown PA, Hanna TN, Juewen BP, Liu, Carter MB, Carroll NJ, Jiang X, Dunphy DR, Willman CL, Petsev DN, Evans DG, Parikh AN, Chackerian B, Wharton W, Peabody DS, Brinker aCJ. The targeted delivery of multicomponent cargos to cancer cells by nanoporous particle-supported lipid bilayers. Nature Materials. 2011;10:389–397. doi: 10.1038/nmat2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graessmann A, Graessmann M, Hoffmann H, Niebel J, Brandner G, Mueller N. Inhibition by interferon of SV40 tumor antigen formation in cells injected with SV40 cRNA transcribed in vitro. FEBS Letters. 1974;39:249–251. doi: 10.1016/0014-5793(74)80122-5. [DOI] [PubMed] [Google Scholar]
- 14.Hochmuth RM. Micropipette aspiration of living cells. J Biomech. 2000;33:15–22. doi: 10.1016/s0021-9290(99)00175-x. [DOI] [PubMed] [Google Scholar]
- 15.Liu XY, Fernandes R, Gertsenstein M, Perumalsamy A, Lai I, Chi M, Moley KH, Greenblatt E, Jurisica I, Casper RF, Sun Y, Jurisicova A. Automated Microinjection of Recombinant BCL-X into Mouse Zygotes Enhances Embryo Development. Plos One. 2011;6(7) doi: 10.1371/journal.pone.0021687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, Kis A, Zettl A, Bertozzi CR. A cell nanoinjector based on carbon nanotubes. Proceeding of the National Academy of Sciences. 2007;104:8218–8222. doi: 10.1073/pnas.0700567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalek AK, Robinson JT, Karp ES, Lee JS, Ahn DR, Yoon MH, Sutton A, Jorgolli M, Gertner RS, Gujral TS, MacBeath G, Yang EG, Park H. Vertical silicon nanowires as a universal platform for delivering biomolecules into living cells. Proceeding of the National Academy of Sciences. 2010;10.1073:1–6. doi: 10.1073/pnas.0909350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singhal R, Orynbayeva Z, Sundaram RVK, Niu JJ, Bhattacharyya S, Vitol EA, Schrlau MG, Papazoglou ES, Friedman G, Gogotsi Y. Multifunctional carbon-nanotube cellular endoscopes. Nature Nanotechnology. 2011;6:57–64. doi: 10.1038/nnano.2010.241. [DOI] [PubMed] [Google Scholar]
- 19.Yum K, Na S, Xiang Y, Wang N, Yu MF. Mechanochemical Delivery and Dynamic Tracking of Fluorescent Quantum Dots in the Cytoplasm and Nucleus of Living Cells. Nano Letters. 2009;9(5):2193–2198. doi: 10.1021/nl901047u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VanDersarl JJ, Xu AM, Melosh NA. Nanostraws for Direct Fluidic Intracellular Access. Nano Letters. 2012;12(8):3881–3886. doi: 10.1021/nl204051v. [DOI] [PubMed] [Google Scholar]
- 21.Almquist BD, Melosh NA. Fusion of biomimetic stealth probes into lipid bilayer cores. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(13):5815–5820. doi: 10.1073/pnas.0909250107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loh O, Lam R, Chen M, Moldovan N, Huang H, Ho D, Espinosa HD. Nanofountain-probe-based high-resolution patterning and single-cell injection of functionalized nanodiamonds. Small. 2009;5(14):1667–1674. doi: 10.1002/smll.200900361. [DOI] [PubMed] [Google Scholar]
- 23.Fu CC, Lee HY, Chen K, Lim TS, Wu HY, Lin PK, Wei PK, Tsao PH, Chang HC, Fann W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proceed Nat Acad Sci. 2007;104:727–732. doi: 10.1073/pnas.0605409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundqvist JA, Sahlin F, Aberg MAI, Stromberg A, Eriksson PS, Orwar O. Altering the biochemical state of individual cultured cells and organelles with ultramicroelectrodes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(18):10356–10360. doi: 10.1073/pnas.95.18.10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ratledge C, Kristiansen B. Basic biotechnology. 3. Cambridge, U.K.; New York: Cambridge University Press; 2006. [Google Scholar]
- 26.Wang MY, Orwar O, Olofsson J, Weber SG. Single-cell electroporation. Analytical and Bioanalytical Chemistry. 2010;397(8):3235–3248. doi: 10.1007/s00216-010-3744-2. [DOI] [PubMed] [Google Scholar]
- 27.Teissie J, Golzio M, Rols MP. Mechanisms of cell membrane electropermeabilization: A minireview of our present (lack of ?) knowledge. Biochimica Et Biophysica Acta-General Subjects. 2005;1724(3):270–280. doi: 10.1016/j.bbagen.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Kim JA, Cho KC, Shin MS, Lee WG, Jung NC, Chung CI, Chang JK. A novel electroporation method using a capillary and wire-type electrode. Biosensors & Bioelectronics. 2008;23(9):1353–1360. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 29.Lee WG, Demirci U, Khademhosseini A. Microscale electroporation: challenges and perspectives for clinical applications. Integrative Biology. 2009;1(3):242–251. doi: 10.1039/b819201d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann E, Schaeferridder M, Wang Y, Hofschneider PH. Gene-Transfer into Mouse Lyoma Cells by Electroporation in High Electric-Fields. Embo Journal. 1982;1(7):841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryttsen F, Farre C, Brennan C, Weber SG, Nolkrantz K, Jardemark K, Chiu DT, Orwar O. Characterization of single-cell electroporation by using patch-clamp and fluorescence microscopy. Biophysical Journal. 2000;79(4):1993–2001. doi: 10.1016/S0006-3495(00)76447-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas K, Sin WC, Javaherian A, Li Z, Cline HT. Single-cell electroporation for gene transfer in vivo. Neuron. 2001;29(3):583–591. doi: 10.1016/s0896-6273(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 33.Haas K, Jensen K, Sin WC, Foa L, Cline HT. Targeted electroporation in Xenopus tadpoles in vivo - from single cells to the entire brain. Differentiation. 2002;70(4–5):148–154. doi: 10.1046/j.1432-0436.2002.700404.x. [DOI] [PubMed] [Google Scholar]
- 34.Nolkrantz K, Farre C, Brederlau A, Karlsson RID, Brennan C, Eriksson PS, Weber SG, Sandberg M, Orwar O. Electroporation of single cells and tissues with an electrolyte-filled capillary. Analytical Chemistry. 2001;73(18):4469–4477. doi: 10.1021/ac010403x. [DOI] [PubMed] [Google Scholar]
- 35.Bestman JE, Ewald RC, Chiu SL, Cline HT. In vivo single-cell electroporation for transfer of DNA and macromolecules. Nature Protocols. 2006;1(3):1267–1272. doi: 10.1038/nprot.2006.186. [DOI] [PubMed] [Google Scholar]
- 36.Judkewitz B, Rizzi M, Kitamura K, Hausser M. Targeted single-cell electroporation of mammalian neurons in vivo. Nature Protocols. 2009;4(6):862–869. doi: 10.1038/nprot.2009.56. [DOI] [PubMed] [Google Scholar]
- 37.Olofsson J, Levin M, Stromberg A, Weber SG, Ryttsen F, Orwar O. Scanning electroporation of selected areas of adherent cell cultures. Analytical Chemistry. 2007;79(12):4410–4418. doi: 10.1021/ac062140i. [DOI] [PubMed] [Google Scholar]
- 38.Rae JL, Levis RA. Single-cell electroporation. Pflugers Archiv-European Journal of Physiology. 2002;443(4):664–670. doi: 10.1007/s00424-001-0753-1. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, Rubinsky B. Microfabricated electroporation chip for single cell membrane permeabilization. Sensors and Actuators a-Physical. 2001;89(3):242–249. [Google Scholar]
- 40.Khine M, Lau A, Ionescu-Zanetti C, Seo J, Lee LP. A single cell electroporation chip. Lab on a Chip. 2005;5(1):38–43. doi: 10.1039/b408352k. [DOI] [PubMed] [Google Scholar]
- 41.Fox MB, Esveld DC, Valero A, Luttge R, Mastwijk HC, Bartels PV, van den Berg A, Boom RM. Electroporation of cells in microfluidic devices: a review. Analytical and Bioanalytical Chemistry. 2006;385(3):474–485. doi: 10.1007/s00216-006-0327-3. [DOI] [PubMed] [Google Scholar]
- 42.Seo J, Ionescu-Zanetti C, Diamond J, Lal R, Lee LP. Integrated multiple patch-clamp array chip via lateral cell trapping junctions. Appl Phys Lett. 2004;84:1973. [Google Scholar]
- 43.Lin YC, Li M, Wu CC. Simulation and experimental demonstration of the electric field assisted electroporation microchip for in vitro gene delivery enhancement. Lab on a Chip. 2004;4(2):104–108. doi: 10.1039/b312804k. [DOI] [PubMed] [Google Scholar]
- 44.Lu H, Schmidt MA, Jensen KF. A microfluidic electroporation device for cell lysis. Lab on a Chip. 2005;5(1):23–29. doi: 10.1039/b406205a. [DOI] [PubMed] [Google Scholar]
- 45.Shin YS, Cho K, Kim JK, Lim SH, Park CH, Lee KB, Park Y, Chung C, Han DC, Chang JK. Electrotransfection of mammalian cells using microchannel-type electroporation chip. Analytical Chemistry. 2004;76(23):7045–7052. doi: 10.1021/ac0496291. [DOI] [PubMed] [Google Scholar]
- 46.Boukany PE, Morss A, Liao WC, Henslee B, Jung HC, Zhang XL, Yu B, Wang XM, Wu Y, Li L, Gao KL, Hu X, Zhao X, Hemminger O, Lu W, Lafyatis GP, Lee LJ. Nanochannel electroporation delivers precise amounts of biomolecules into living cells. Nature Nanotechnology. 2011;6(11):747–754. doi: 10.1038/nnano.2011.164. [DOI] [PubMed] [Google Scholar]
- 47.Wang JP, Yang SC, Wang C, Wu QO, Wang ZY. A DEP-Assisted Single-Cell Electroporation Chip with Low Operation Voltage. 2010 Ieee Sensors. 2010:2097–2100. [Google Scholar]
- 48.Odorizzi L, Ress C, Collini C, Morganti E, Lorenzelli L, Coppedè N, Alabi AB, Iannotta S, Cazzanelli E, Vidalino L, Macchi P. An integrated platform for in vitro single-site cell electroporation: Controlled delivery and electrodes functionalization. Sensors and Actuators B: Chemical. (0) [Google Scholar]
- 49.Valero A, Post JN, van Nieuwkasteele JW, ter Braak PM, Kruijer W, van den Berg A. Gene transfer and protein dynamics in stem cells using single cell electroporation in a microfluidic device. Lab on a Chip. 2008;8(1):62–67. doi: 10.1039/b713420g. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Stine MJ, Lu C. Microfluidic cell electroporation using a mechanical valve. Analytical Chemistry. 2007;79(24):9584–9587. doi: 10.1021/ac701854b. [DOI] [PubMed] [Google Scholar]
- 51.Fei ZZ, Hu X, Choi HW, Wang SN, Farson D, Lee LJ. Micronozzle Array Enhanced Sandwich Electroporation of Embryonic Stem Cells. Analytical Chemistry. 2010;82(1):353–358. doi: 10.1021/ac902041h. [DOI] [PubMed] [Google Scholar]
- 52.Khine M, Ionescu-Zanetti C, Blatz A, Wang LP, Lee LP. Single-cell electroporation arrays with real-time monitoring and feedback control. Lab on a Chip. 2007;7(4):457–462. doi: 10.1039/b614356c. [DOI] [PubMed] [Google Scholar]
- 53.Xie C, Lin ZL, Hanson L, Cui Y, Cui BX. Intracellular recording of action potentials by nanopillar electroporation. Nature Nanotechnology. 2012;7(3):185–190. doi: 10.1038/nnano.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nawarathna D, Unal K, Wickramasinghe HK. Localized electroporation and molecular delivery into single living cells by atomic force microscopy. Applied Physics Letters. 2008;93(15) [Google Scholar]
- 55.Moldovan N, Kim KH, Espinosa HD. Design and fabrication of a novel microfluidic nanoprobe. J MEMS. 2006;15(1):204–213. [Google Scholar]
- 56.Loh O, Ho A, Rim J, Kohli P, Patankar N, Espinosa H. Electric field-induced direct delivery of proteins by a nanofountain probe. PNAS. 2008;105(43):16438–16443. doi: 10.1073/pnas.0806651105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim KH, Sanedrin RG, Ho AM, Lee SW, Moldovan N, Mirkin CA, Espinosa HD. Direct delivery and submicrometer Patterning of DNA by a nanofountain probe. Advanced Materials. 2008;20(2):330. [Google Scholar]
- 58.Wu B, Ho A, Moldovan N, Espinosa HD. Direct deposition and assembly of gold colloidal particles using a nanofountain probe. Langmuir. 2007;23(17):9120–9123. doi: 10.1021/la7011952. [DOI] [PubMed] [Google Scholar]
- 59.Krassowska W, Filev PD. Modeling electroporation in a single cell. Biophysical Journal. 2007;92(2):404–417. doi: 10.1529/biophysj.106.094235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agarwal A, Zudans I, Weber EA, Olofsson J, Orwar O, Weber SG. Effect of cell size and shape on single-cell electroporation. Analytical Chemistry. 2007;79(10):3589–3596. doi: 10.1021/ac062049e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kotnik T, Bobanovic F, Miklavcic D. Sensitivity of transmembrane voltage induced by applied electric fields - a theoretical analysis. Bioelectrochemistry and Bioenergetics. 1997;43(2):285–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.