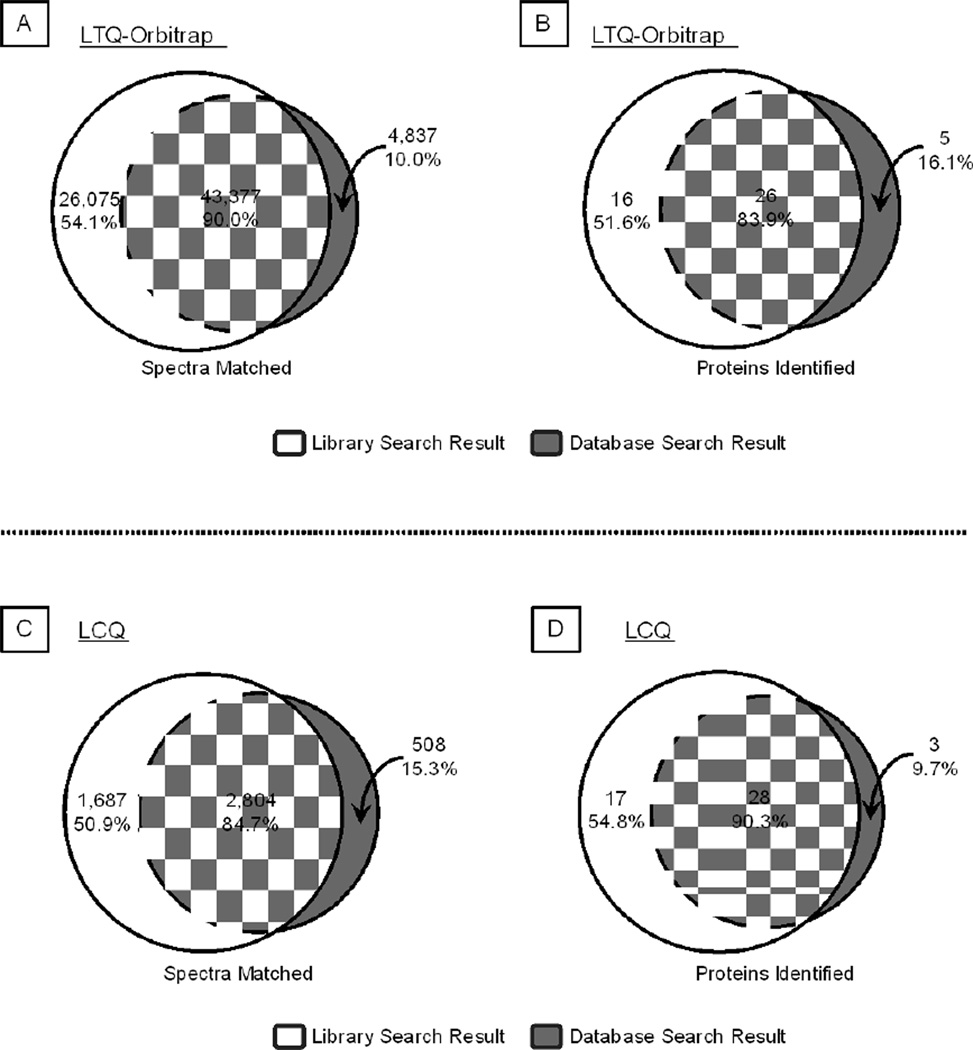
Figure 5.
Benchmark Tests of Library Search Engine in Protein Identification. A–B. With the test dataset of murine 20S proteasome spectra collected by an LTQ-Orbitrap mass spectrometer, the library search workflow covered 90.0% of the spectral matches (A) and 83.9% of protein identifications (B) provided by a sequence database search workflow. Furthermore, the library search offered an additional 54.1% unique spectral matches and 51.6% unique protein identifications. C–D. With the LCQ test dataset, the library search workflow covered 84.7% of spectral matches (C) and 90.3% of protein identifications (D) that were captured via a sequence database search workflow. The library search also offered an additional 50.9% unique spectral matches and 54.8% unique protein identifications.