Figure 1. Types of protein complexes designed using computational methods.
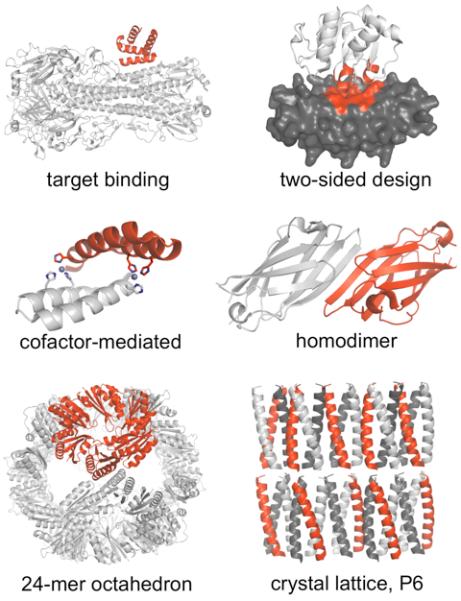
Target binding [32]: a helical scaffold (red) redesigned to bind the stem region of influenza hemagglutinin (white). Two-sided design [48]: redesigned scaffolds were ankyrin repeat protein (gray) and a coenzyme A binding protein (PH1109, white), interface contacts in red. Cofactor-mediated binding [39]: A helical hairpin designed for zinc-mediated homodimerization. Histidine residues (sticks) coordinate zinc (spheres). Homodimer [38]: the γ-adaptin appendage domain – a monomer with an exposed beta strand – redesigned to allow intermolecular beta-sheet formation. Nanohedra [17]: a native trimer (red) redesigned to form an octamer of trimers, a 24-mer octahedron. P6 crystal lattice [45]: a previously designed coiled-coil homotrimer modified to form a predetermined crystal lattice in a rare space group, P6.
