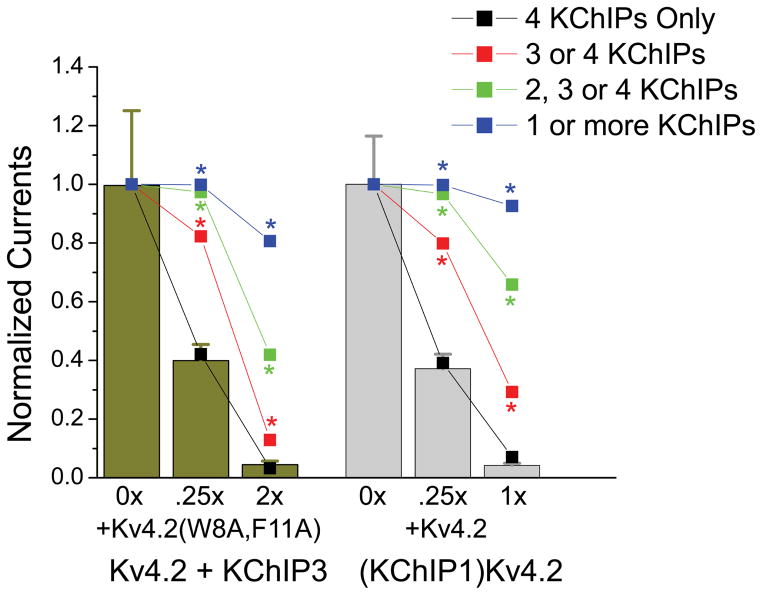
Figure 8. Binomial assembly modeling shows that titration experiment results are predicted if 4 KChIP proteins must be bound to the channel to enhanced functional expression.
Results from our titration experiments presented in Figs. 3 and 6 were compared to model predictions based on the need to incorporate 1, 2, 3, or 4 KChIPs into a channel in order to enhance functional expression. Results show that only the model where 4 KChIPs must be bound to enhance functional expression provides an accurate fit with predictions under all conditions tested that are not significantly different from our data. This result is consistent with a single unbound FERN domain fully suppressing enhanced channel functional expression. All other model predictions are significantly different from the measured data under the conditions indicated by the asterisks.