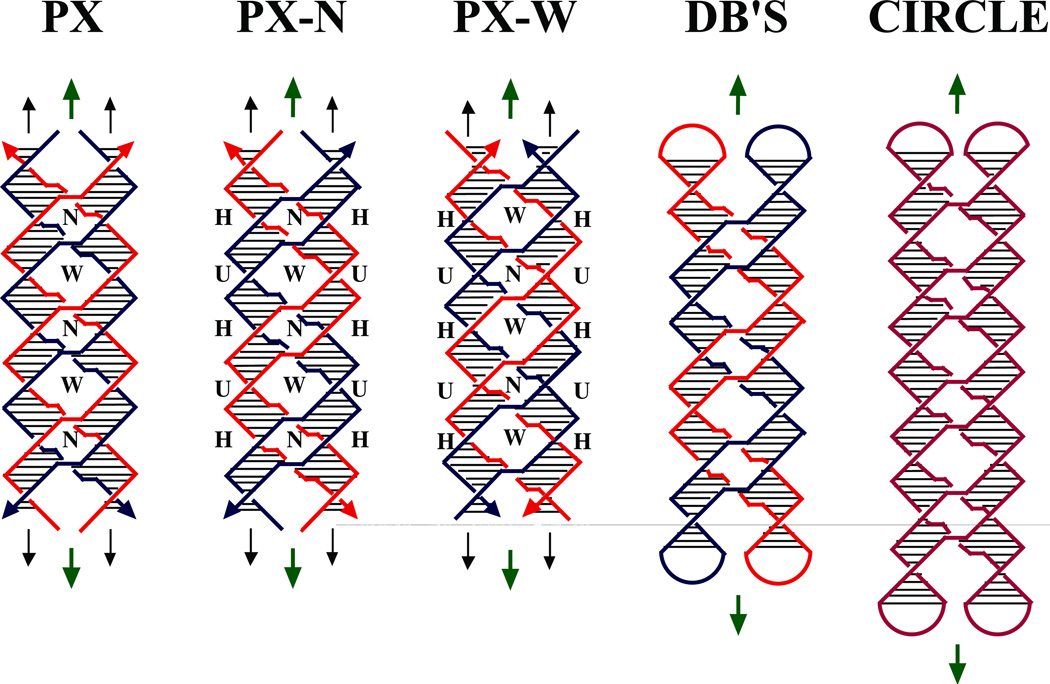
Figure 1. Schematic Drawings of Paranemic Crossover DNA, and its Closed Analogs.
The three molecules on the left are drawn containing pairs of strands drawn in red and blue; strands drawn with the same color are related to each other by the dyad axis. The helix of each strand is approximated by a zig-zag structure. Arrowheads on the strands denote their 3' ends. The base pairs are indicated by very thin black lines. Both the PX and PX-N molecules contain four strands, arranged in two double helical domains related by a central dyad axis. The PX and PX-N molecules are identical, but two different pairs of dyad symmetries are shown between strands flanking the dyad: PX illustrates symmetry between strands of the same polarity, whereas PX-N shows symmetry between strands flanking a minor groove (a third symmetry, between strands flanking the major groove, is not shown). The view is perpendicular to the plane containing both helix axes. The dyad axes are indicated by the short green arrows above and below each molecule. The PX and PX-N molecules have alternating major (wide) and minor (narrow) groove tangles, indicated by 'W' and 'N', respectively. The formation of PX from two intact double helices would require homology in the unit tangles labeled 'H', but not those labeled 'U'. The two molecules on the far right are closed PX molecules. The left molecule of the two contains two paired dumbbells (DB's) that are unlinked topologically. The molecule on the right is a half-turn longer in each helical domain; the resulting structure is an intricately self-paired single-stranded circle drawn in purple.