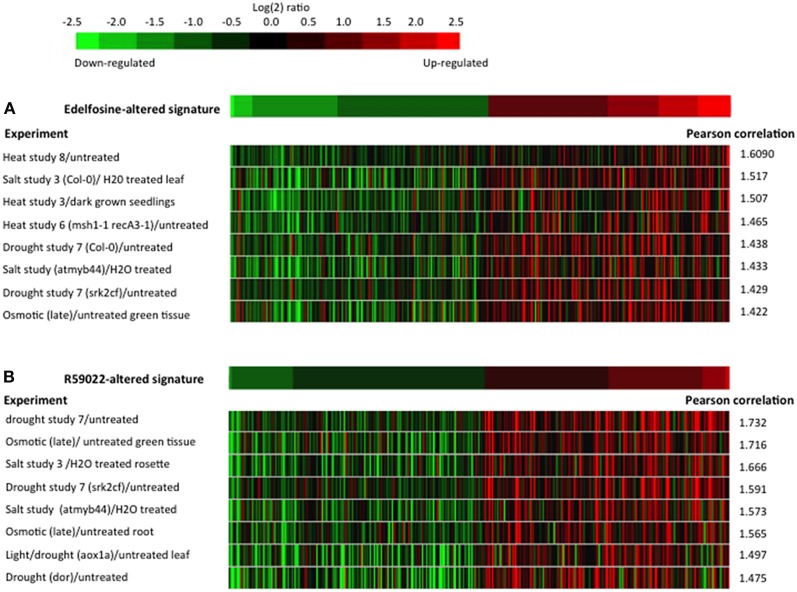
Figure 5.
Similarity between the edelfosine-responsive or R59022-responsive transcriptomes and the stress-responsive transcriptomes. (A) The 200 genes that are most up-regulated by edelfosine and the 200 genes that are most down-regulated by edelfosine were used as a signature to search for the transcriptome experiments with the highest similarity. (B) The 200 genes that are most up-regulated by R59022 and the 200 genes that are most down-regulated by R59022 were used as a signature to search for the transcriptome experiments with the highest similarity. The similarity search was performed against the 789 experiments classified as “stress” by Genevestigator (Hruz et al., 2008). The experiments are sorted according to the Pearson's correlation. The expressions of the signature genes in the 8 most similar experiments are shown in color-scale. msh1-1recA3-1, atmyb44, srk2cf, aox1, dor are mutant plants in MSH1 and RecA3, MYB transcription factor 44 (Jung et al., 2008), SnRK2C and SnRK2F (Mizoguchi et al., 2010), alternative oxidase1 (Giraud et al., 2008), and the F-box protein DOR (Zhang et al., 2008), respectively.