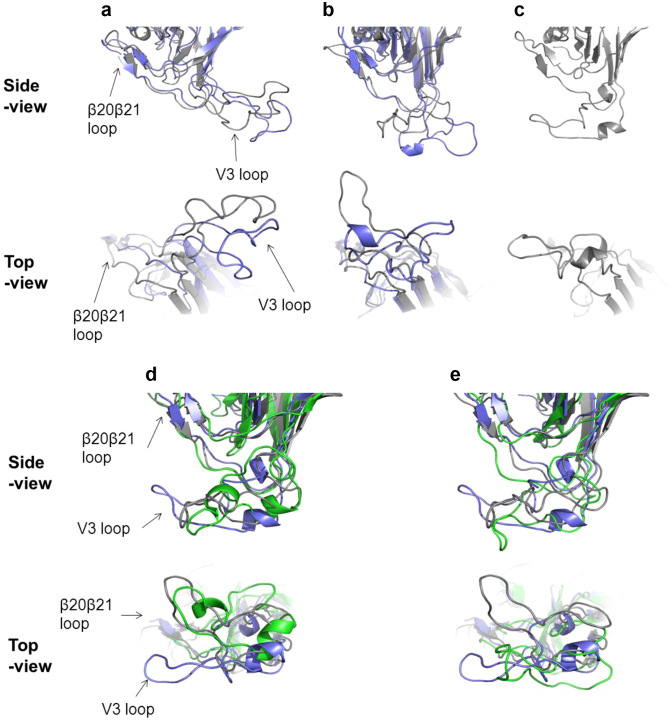
Figure 3.
Structural models of V3 loops on HIV-1 gp120 outer domains (a, b and c). MD simulations were performed for the HIV-1 JRFL gp120 outer domain with various V3 loops for CCR5 (a), dual (b), and CXCR4 (c) tropism. The most frequently appeared structures during 5–10 ns of MD simulations were extracted, and the top and side views of the structures around V3 loops are highlighted. (a) JRFL (gray) and Del25 (navy). (b) Ins11R/S12H/Del25 (gray) and Ins11R/S12H (navy). (c) CL8 (gray). Structural models of V3 loops of Ins11R/S12H/Del25-derived mutants (d and e). MD simulations were performed for the HIV-1 Ins11R/S12H/Del25 gp120 outer domain with D29N (d) or T8V (e) substitution in V3 loop. The most frequently appeared structures during 5–10 ns of MD simulations were extracted and superimposed with those of Ins11R/S12H/Del25 and CL8. (d) Superimposition of D29N (green), Ins11R/S12H/Del25 (gray), and CL8 (navy). (e) Superimposition of Ins11R/S12H/Del25/T8V (green), Ins11R/S12H/Del25 (gray), and CL8 (navy). Top and side views of the structures around V3 loops are shown.