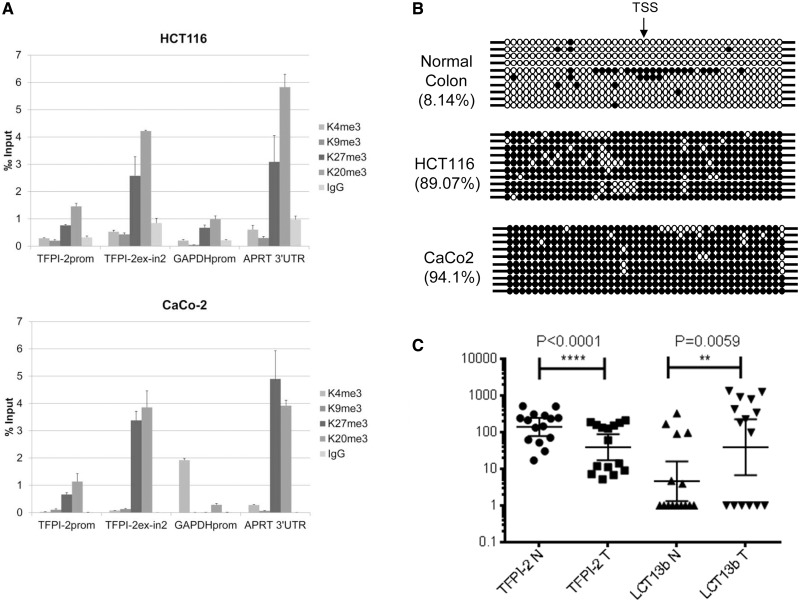
Figure 7.
Silencing of TFPI-2 in colorectal cancer. (A) Chromatin immunoprecipitation assays in HCT116 (top) and CaCo-2 (bottom) cells using antibodies against active histone marks AcH3, H3K4me3, and repressive histone marks H3K9me3, H3K27me3, and H4K20me3 indicate enrichment of repressive marks H3K27me3 and H4K20me3 at the TFPI-2 regions tested (see Figure 5A). (B) Bisulphite sequencing analysis of 10 independent clones from normal colon and HCT116 and CaCo-2 colon cancer cells shows that the TFPI-2 region studied is hypomethylated in normal colon (8.14% methylation) and hypermethylated in HCT116 and CaCo-2 cells (89.07 and 94.1% methylation, respectively). TSS indicates the transcriptional start site of the TFPI-2 gene; filled and open circles represent methylated and unmethylated CpG dinucleotides, respectively. Each row represents an independent clone. The region analysed by bisulphite sequencing is depicted in Figure 5A. (C) Analysis of LCT13 and TFPI-2 expression in matched normal and tumour tissues from sporadic cases of colorectal cancer shows an inverse relationship with a significant increase in LCT13 (P < 0.007, Wilcoxon test, n = 15) corresponding to a significant decrease in TFPI-2 in tumours. Real-time RT–PCR expression data was normalized to PGK1, GAPDH, and HPRT using GenEx software (Multid Analyses AB).