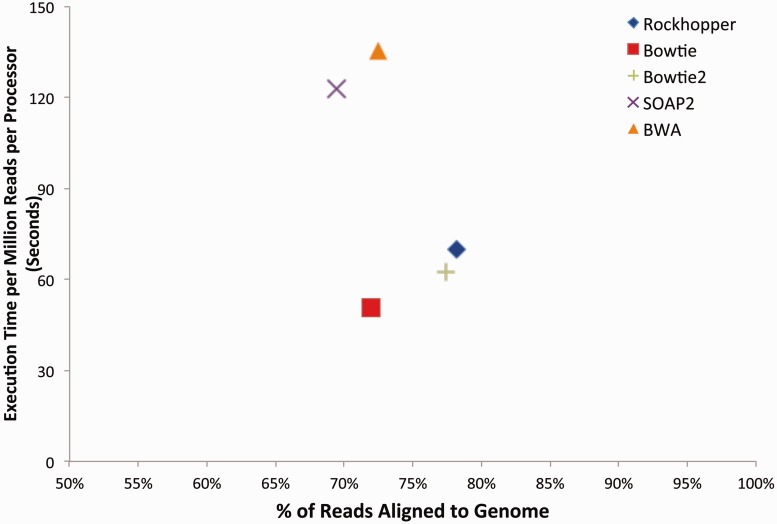
Figure 2.
Aligning sequencing reads to a genome. The performance of five tools for aligning reads to a genome is shown. The five tools are Rockhopper (version 1.00), Bowtie (version 0.12.7), Bowtie2 (version 2.0.0-beta5), SOAP2 (version 2.21) and BWA (version 0.6.2). Each tool is based on an FM-index, and each tool was executed on the same machine with default parameters using the same number of processors. The tools were evaluated by the percentage of 2 134 636 656 reads that they successfully aligned to a reference genome (x-axis) and by the execution time they required per million reads per processor (y-axis). The reads come from 75 RNA-seq experiments conducted using five different bacteria.