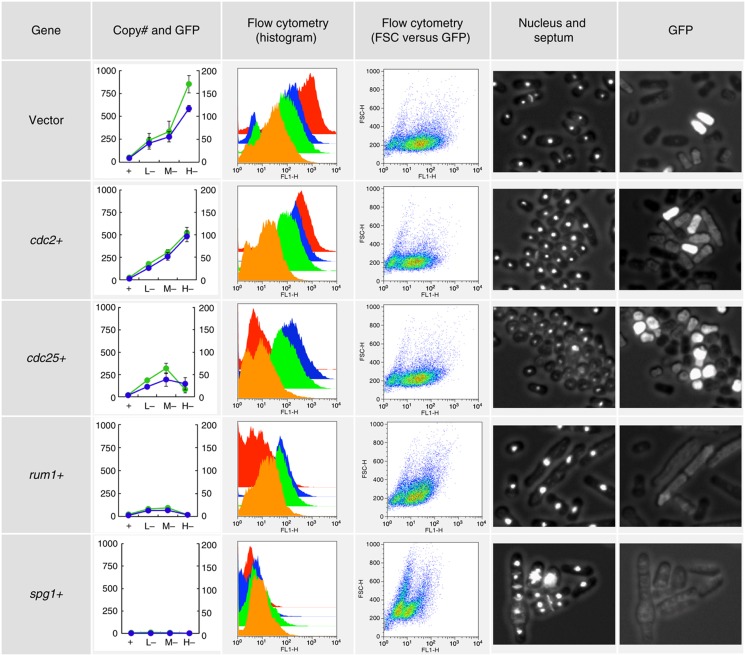
Figure 2.
Examples of gTOW experiments with cdc genes in S. pombe. The experimental results obtained by gTOW with empty vector, cdc2+, cdc25+, rum1+, and spg1+ are shown (enlarged part of figure is shown in Supplementary Figure S5). First column: The plasmid copy number (blue graph, right vertical axis) and the mean GFP fluorescence (green graph, left vertical axis) in gTOW in leucine+ (indicated as ‘+’) and leucine− with each vector (pTOWsp-L, -M, and -H) are shown (indicated as ‘L−,’ ‘M−,’ and ‘H−,’ respectively). The original data are shown in Supplementary Tables S6 and S7. Second column: Cell distribution with GFP fluorescence in gTOW experiments in leucine+ (orange graph) and leucine− conditions with −L vector (green graph), −M vector (blue graph), and −H vector (red graph) are shown. Some experiments showed bimodality distributions. This is probably due to the plasmid loss, which is a property of the ars-based plasmid. Third column: Scatter plot between GFP fluorescence (FL-1) and cell size (FSC). Cells were cultured in the leucine+ condition. Fourth column: Fluorescent microscopic image of the cells cultured in the leucine+ condition. Nucleus and septum are stained. Fifth column: GFP fluorescence of the cells in the fourth column. The brightness reflects the plasmid copy number within the cell. The whole data set of above analysis for the genes analyzed in this study is provided upon request.