Figure 6.
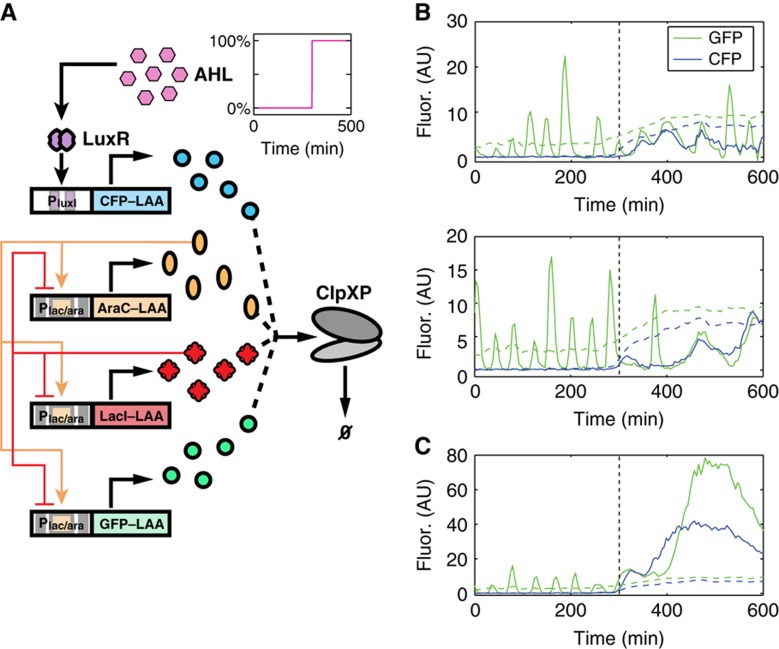
Coupled degradation can serve to indirectly couple gene circuits. (A) Circuit diagram for a variant of a synthetic gene oscillator discussed previously (Stricker et al, 2008), expressed alongside an AHL-inducible, LAA-tagged CFP (see Supplementary Information for details). Competition for enzymatic degradation among LAA-tagged proteins allows CFP to interact with the oscillator by slowing the degradation of the oscillator components. Since fast enzymatic decay is thought to strongly influence the period and robustness of certain gene oscillators (Wong et al, 2007; Mather et al, 2009), the character of GFP oscillations should be closely tied to CFP expression. (B) Cells containing the circuit in (A) were grown in a microfluidic device to test the influence of CFP production on GFP oscillations (see Supplementary Information for details). Two single-cell trajectories for GFP and CFP fluorescence (solid lines) show regular oscillations in GFP fluorescence in the absence of external AHL, that is, at low CFP fluorescence. However, addition of 15 nm AHL (time of induction start is indicated by a vertical dotted line) introduces CFP into the system, causing the GFP oscillations to slow and the CFP signal to oscillate as a result of indirect coupling due to queueing. Coupling is also observed in the mean fluorescence across a region of cells (mean fluorescence as dashed lines), where increasing mean CFP fluorescence is associated with an increase in mean GFP fluorescence. (C) A representative trajectory where both CFP and GFP become and remain bright simultaneously. Positive coupling between trajectories is apparent.