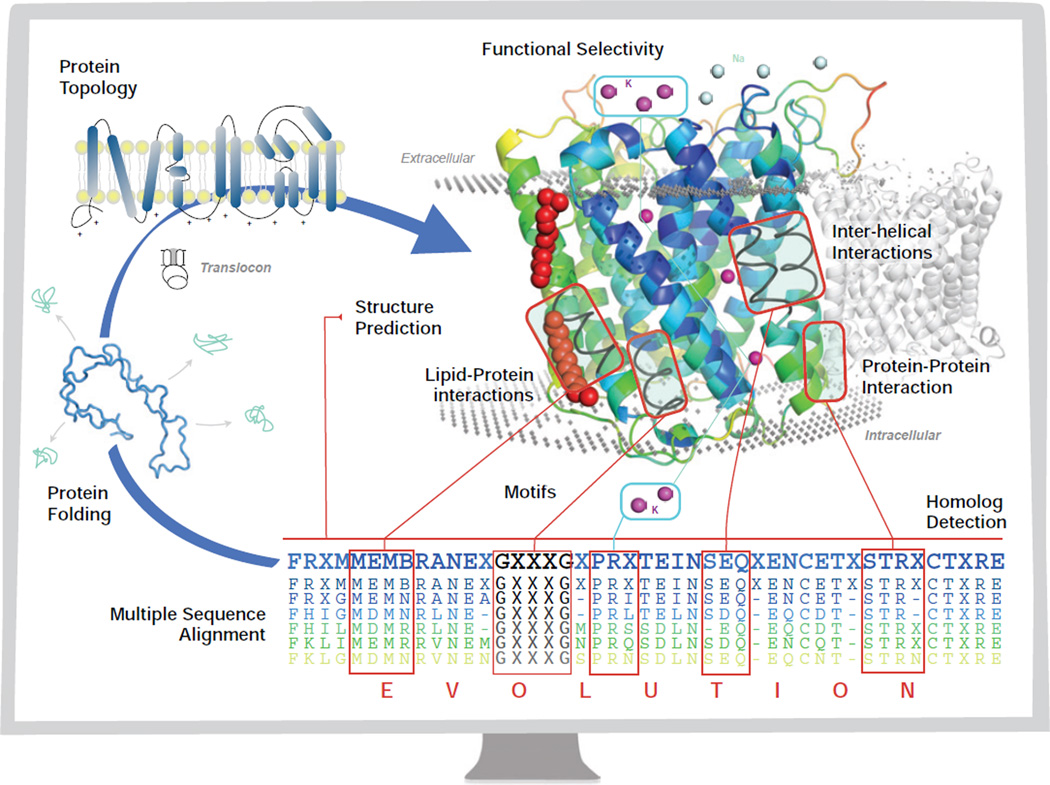
Figure 1.
The central dogma of molecular biology for membrane proteins (blue arrow) through the monitor of a computer. The chain of amino acids folds through the mediation of the translocon to a final stable low energy structure, with a specific topology (Section 2.2). The goal of structure prediction is to derive the three-dimensional structure of a membrane protein from its sequence (Section 7). The assembly of the helices in the transmembrane domains is facilitated by interhelical interactions (Section 6) via sequence and spatial motifs (Section 3), as well as protein lipid interactions (Sections 5.2 and 4). Membrane proteins also participate in protein-protein interactions (Section 8.2) for biological functions. The evolutionary relationship between membrane proteins can be detected through (multiple) sequence alignment, for which an evolutionary model of substitutions of residues in the transmembrane domain is essential in deriving specialized scoring matrices for alignment and for detection of homologs (Section 4). With significant understanding of the organizing principles of membrane proteins, computational studies can be carried out to design membrane proteins with desired properties such as functional selectivity (Section 8.3).