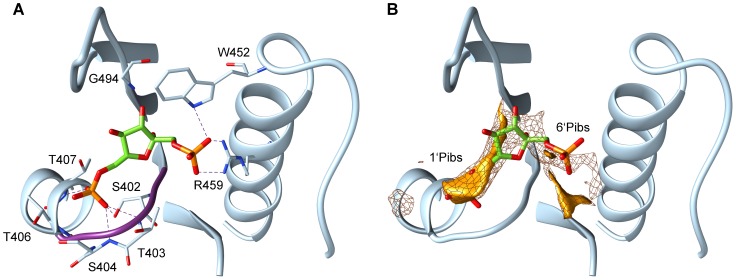
Figure 1. Allosteric binding site of PYK with the activator FBP bound.
Residues V400 to V411, K446 to F470 and S481 to Q496 of the crystallographically resolved Saccharomyces cerevisiae PYK (PDB id: 1A3W, chain A) are shown in cartoon representation. Panel A shows possible hydrogen bonds between the FBP phosphate groups and the residues of PYK within a distance of 3 Å (all in stick representation). The structural P-loop motif (STSG) is coloured in purple. Panel B illustrates the phosphate interaction sites computed with the GRID program. The binding site of the 1′-phosphate moiety of FBP in the allosteric site is referred to as 1′Pibs and that of the 6′-phosphate moiety of FBP as 6′Pibs. The interaction energy is displayed at isocontours of −10 kcal/mol (mesh surface) and −13.5 kcal/mol (solid surface). Only the phosphate interaction sites that are located within 6 Å around FBP are shown.