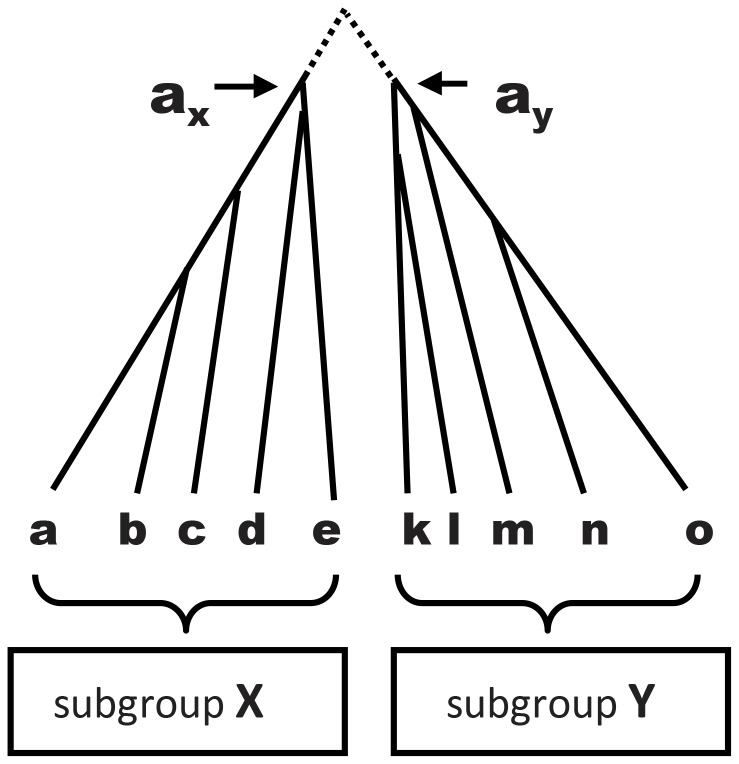
Figure 1. We use two natural subgroups (X and Y), independently align the sequences for the species in each subgroup, independently determine the optimal tree for each subgroup, independently infer the ancestral sequences ax and ay on the optimal subtrees (in practice the sequence at the nearest node to the root of the subtree is estimated), and finally measure the pairwise alignment score between the ancestral sequences, s(ax,ay).
Separately, we measure the alignment score between each pair of sequences (s(i,j)) with one member in each of the two subsets, for example, s(a,k), s(a,l), s(a,m), and so on.