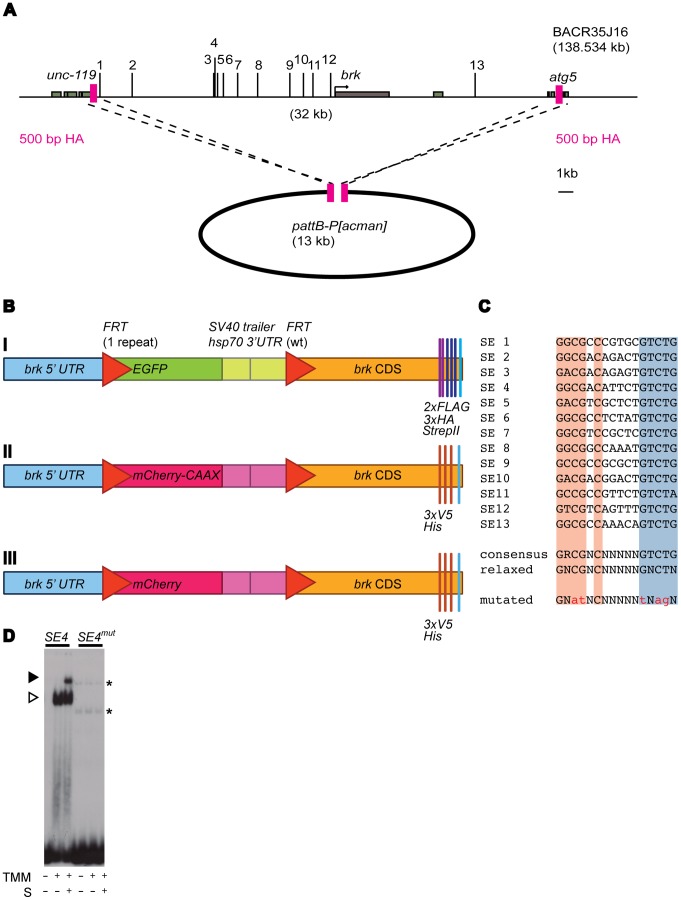
Figure 1. Generation of tagged and fluorescently labeled genomic brk constructs.
(A) Homologous recombination mediated retrieval of the genomic region of interest into the integration vector pattB-P[acman]. Schematic overview of the brk locus, the distribution of the 13 putative SEs is indicated. The brk coding sequence and the up- and downstream neighboring genes (unc-119 and atg5, respectively) are shown in gray, 500 bp homology arms (HA) in pink. Fragment B14 from Muller et al. contains SEs 3–8 [14]. SEs 11–2 from Yao et al. correspond to SEs 1–10 in this current study; SE 1 corresponds to SE 12 in this current study [15]. (B) Schematic overview of the EGFP and mCherry flip out cassettes as well as the C-terminal tags. (I) The EGFP flip out cassette and C-terminal FLAG-, HA-tags and strep-tagII that were introduced in the 10 constructs that were generated in this work, featuring from zero to 13 functional SEs. (II) A membrane-targeted mCherry in a flip out cassette and C-terminal V5- and His-tags were used in the wild type construct that served as an internal control for the quantification. (III) Construct similar to the one above, except that the mCherry in this construct lacks the CAAX motif and is hence no longer recruited to the cell membrane (see Text S1). (C) Sequence summary of the 13 putative SEs. The Mad and Med binding motifs are shown in red and blue, respectively. A spacer of five random nucleotides and the T at position 15 allow Shn binding. Shown is the consensus sequence as previously described (“consensus” according to [2], [8]). For this project, we allowed the consensus to become more degenerate, or “relaxed”, allowing more mismatches compared to the original consensus sequence. The five point mutations that were introduced into each SE are shown in lower case (“mutated”). (D) EMSA with SE4 as a representative example to examine whether the introduced mutations prevent complex formation. Results shown for both wild type SE4 and SE4mut. Each labeled DNA was incubated with extracts of mock transfected cells (first lane) or extracts of cells transfected with TkvQD, Mad and Medea (TMM) in the absence (second lane) or presence (third lane) of ShnCT (S). Open arrow: Mad-Med shift, closed arrow: Mad-Med-ShnCT super shift. Nonspecific binding events are indicated by asterisks.