Abstract
Smoothened (SMO) is an important member of the Hedgehog signaling pathway. We constructed a specific recombinant lentiviral vector for RNA interference, targeting the SMO gene (NM_005631) to observe its effect on SMO expression, cell proliferation and the cell cycle in the human androgen-sensitive prostate cancer cell line, LNCaP, and in the androgen-independent prostate cancer cell line, PC3. Four siRNA sequences were designed and inserted into a lentiviral vector pGCSIL-GFP to construct four recombinant vectors. The vector with the highest interfering efficiency was co-transfected with packaging vectors (pHelper1.0 and pHelper2.0) in 293T cells to assemble lentivirus particles by liposome for infecting LNCaP and PC3 cell lines, respectively. The expression level of SMO mRNA, tumor cell proliferation and cell cycle were measured by quantitative realtime polymerase chain reaction (qRT-PCR), 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide (MTT) assay and flow cytometry, respectively. Sequence results showed that recombinant lentiviral vectors were constructed successfully. pGCSIL-GFP-723 had the highest interfering efficiency, named Lv-SIL-SMO723 after co-transfection, with which LNCaP and PC3 cell lines were infected. Compared with the control groups, results showed significantly decreased (P < 0.05) SMO mRNA expressions of LNCaP and PC3, lower mean percentage of S-phase cells and higher mean percentage of G2/M phase cells, as well as obviously slow proliferation (P < 0.01) of LNCaP in the infected group. Yet, the proliferation of PC3 was not altered (P > 0.05). In conclusion, the recombinant lentivirus particles were able to suppress SMO expression, regulate the cell cycle in the LNCaP and PC3 cell lines and markedly inhibit proliferation of LNCaP cells but not PC3 cells.
Keywords: lentivirus, prostate neoplasm, RNAi, smoothened
Introduction
Prostate cancer is one of the most common cancer in men. Studies show that the occurrence and development of prostate cancer is relevant to the activation of the Hedgehog (Hh) signaling pathway 1. The proto-oncogene Smoothened (SMO) receptor is one of the key components of Hh receptors. SMO protein, as one of the transmembrane proteins, activates the Hh signaling pathway. Furthermore, the expression of the SMO gene occurs only in prostate cancer tissues but not in benign prostate epithelial cells 2. Targeted inhibition of the SMO gene in controlling activities of the Hh signaling pathway has a critical significance in prostate cancer therapy 3. This study constructs recombinant lentivirus particles for RNA interference (RNAi) that targets the SMO gene by co-transfecting 293T cell lines with a lentiviral vector containing green fluorescent protein (GFP). The recombinant lentivirus particles with high interfering efficiency and stable gene expression were used to infect the human androgen-sensitive prostate cancer cell line, LNCaP, and the androgen-independent prostate cancer cell line, PC3. The SMO expression, cell proliferation and the cell cycle after the infection were observed and the application of SMO in the treatment of prostate cancer was discussed.
Materials and methods
Materials
The LNCaP, PC3, and 293T cell lines (for lentiviral packaging) were purchased from the gene bank at the Shanghai Institute of Biology, Chinese Academy of Sciences. Dulbecco's Modified Eagle Media (DMEM), fetal bovine serum, trypsin and polymerase chain reaction (Opti-MEM) were purchased from GIBCO (Carlsbad, CA, USA). The Escherichia coli strain DH5α, the lentiviral vector pGCSIL-GFP, packaging system pHelper1.0 and pHelper2.0, virus particles (for the negative control group) and primer synthesis were provided by Shanghai GeneChem Co. Ltd., (Shanghai, China). The AgeI, EcoRI, restriction enzyme HindIII, T4 DNA ligase and T4 DNA ligase buffer were provided by NEB. The Plasmid DNA Extraction (Mini) Kit was provided by QIAGEN (Crawley, West Sussex, UK). Trizol and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA, USA). The SYBR Master Mixture was purchased from TAKARA (Otsu, Shiga, Japan). The Total RNA Isolation Kit, M2MLV Reverse Transcriptase, MgCl2, dNTP, RNase Inhibitor and PGEM2T were purchased from Promega (Madison, WI, USA). Oligo dT was purchased from Shanghai Sangon Company (Shanghai, China). The primary antibody rabbit anti-human Smoothened was purchased from Upstate. MTT was purchased from Beijing Ding Guo BioTech Company. The single stain cell cycle detection kit (PI) was purchased from KeyGEN (Nanjing, China). Sequencing reaction was detected by an ABI 3733 DNA sequencing machine (Majorbio).
Construction of lentiviral and eukaryotic expression vectors and screening of target sites
On the basis of GenBank information for SMO (NM_005631), we designed four small-interfering RNA (siRNA) sequences:
PscSI721 SMO human: AGCGGATCAAGAA-GAGCAA.
PscSI722 SMO human: ATCGCTACCCTGC-TGTTAT.
PscSI723 SMO human: CTCTGTCCTGCGT-CATCAT.
PscSI724 SMO human: CTGTGGCAATCCT-TGCTGT.
We designed DNA oligonucleotides and introduced AgeI and EcoRI restriction sites at the end. A double-stranded DNA oligonucleotide, annealed from two single-stranded ones, was connected to the lentiviral vector pGCSIL-GFP, which was double digested with AgeI and EcoRI. Then, DH5α-competent E. coli cells were transformed. We then selected the colony for polymerase chain reaction (PCR), and the recombinant plasmids were extracted for sequence detection. The lentiviral vector pGCSIL-GFP (GeneChem) was used to construct SMO RNAi lentiviral expression vectors pGCSIL-GFP-SMO721, pGCSIL-GFP-SMO722, pGCSIL-GFP-SMO723 and pGCSIL-GFP-SMO724. The lentiviral vector pGCSIL-GFP-Negative (GeneChem), as a negative control, was used to monitor nonspecific responses caused by heterologous siRNA. We removed the endotoxin in both the SMO RNAi lentiviral expression vectors and pGCSIL-GFP negative and extracted plasmids.
To construct the eukaryotic expression vector, we used a clone containing SMO gene (NM_005631) provided by Life Science College (Tongji University, Shanghai, China). On the basis of the gene information, we designed the primers SMO-HindIII: CCCAAGCTTATGGCCGCTGCCCGCCC and SMO-AgeI: GGCGACCGGTGGGAAGTCCGAGTCTGCATCCATGAG. A full-length sequence of SMO gene, obtained by PCR, and pEGFP-N1 were, respectively, double digested with HindIII and AgeI. After purification, the product of the enzyme digestions was connected directionally and DH5α-competent E.coli cells were transformed. The colony was selected for PCR, and then the recombinant plasmids were extracted for sequence detection. Lastly, the endotoxin was removed from the recombinant eukaryotic expression vector (pEGFP-N1-SMO) and the plasmid was extracted.
Using the instructions for Lipofectamine 2000 (Invitrogen), we co-transfected the extracted recombinant lentiviral expression vector plasmids and the pEGFP-N1-SMO in cultured 293T cell lines. After 24 h, we observed the transfection effect under a fluorescent microscope. After 36 h, we harvested the cells, extracted the protein and used Western blot testing to analyze it. We then identified the target site with the highest interfering efficiency.
Lentiviral packaging and titering
We packaged the recombinant lentiviral vector for SMO RNAi with the highest interfering efficiency. After removing the endotoxins from SMO RNAi lentiviral expression vectors and the packaging vectors (pHelper1.0 and pHelper2.0, respectively), we co-transfected the three types of vectors in 293T cell lines, according to the instructions for Lipofectamine 2000. At 8 h after co-transfection, the cells were transferred to a complete medium. After 48 h, lentivirus supernatant was harvested and concentrated. The virus titer was determined and calibrated in the 293T cell lines. The 293T cell culture medium was then centrifuged and concentrated to obtain an SMO siRNA lentivirus solution, which was named Lv-SIL-SMO723 and kept in a collection cup at −80°C.
The concentrated virus stock was diluted 10 × and then used to transfect 293T cells. To obtain the titer of virus stock, we multiplied the number of fluorescent cells in 10% of the well and the corresponding dilution fold.
SMO RNAi lentivirus particles transfect LNCaP and PC3 cells
We used three parallel wells for each group of cells: non-transfected LNCaP and PC3 cells (control group), pGCSIL-GFP-Negative-transfected LNCaP and PC3 cells (negative control group) and Lv-SIL-SMO723-transfected LNCaP and PC3 cells (knockdown group). LNCaP and PC3 cells for each group were seeded in a six-well plate and incubated at 37°C in 5% CO2 until cell confluence reached 30%. Later we added 2 × 106 TU per well of the recombinant lentivirus to the plate and cultured it in the incubator at 37°C in 5% CO2 for 3 days.
Assay of transfection efficiency
At 3 days after the transfection, we used the FACSCalibur (BD, Franklin Lakes, NJ, USA) flow cytometer to detect transfection efficiency.
qRT-PCR detection of SMO mRNA expression in LNCaP and PC3 cells
At 5 days after the transfection, we extracted the total RNA from cells harvested from the different groups. First-strand cDNA was synthesized by reverse transcription according to the instructions for M2MLV (Promega). For SMO, we used upstream primer GGCTGCTGAGTGAGAAG and downstream primer CTGGTTGAAGAAGTCGTAGAAG. For the internal reference GAPDH, we used upstream primer GGCGGCACCACCATGTACCCT and downstream primer AGGGGCCGGACTCGTCATACT. We used the SYBR Master Mixture (TAKARA, Otsu, Shiga, Japan) to perform the qRT-PCR and the 2−ΔΔCt method for data analysis. The reaction cycle conditions were as follows:
Cycle 1: (1 ×) Step 1: 95.0°C for 15 min.
Cycle 2: (45 ×) Step 1: 95.0°C for 5 min. Step 2: 60.0°C for 30 min.
Cycle 3: (1 ×) Step 1: 95.0°C for 15 min. Step 2: 60.0°C for 1 h. Step 3: 95.0°C for 15 min.
Cell proliferation assay
At 5 days after the transfection, the cell suspension was diluted to 2 × 104 mL−1. Each well of three 96-well culture plates (one plate for each group) was inoculated with a 100-μL cell suspension. The culture was stopped on days 1, 2, 3, 4 and 5. At 4 h before stopping the culture, 10 μL of thiazolyl blue (5 mg mL−1) was added to each well. We carefully aspirated the culture supernatant and then added 150 μL of dimethyl sulfoxide to each well. After 10 min of shaking, we assessed the absorbance value (A value) at 570 nm.
Cell cycle analysis
At 5 days after the transfection, according to the instructions from the kit, we used FACSCalibur (BD, Franklin Lakes, NJ, USA) flow cytometer to assess cell cycles for each group.
Statistical analysis
We used SPSS12.0 (SPSS Inc, Chicago, IL, USA), ANOVA and paired t-test to analyze data. P < 0.05 was considered significant.
Results
Sequencing results
Sequencing results show successful constructions of four recombinant lentiviral vectors (pGCSIL-GFP-SMO721, pGCSIL-GFP-SMO722, pGCSIL-GFP-SMO723 and pGCSIL-GFP-SMO724), and of the eukaryotic expression vector pEGFP-N1-SMO. After co-transfection in 293T cells, Western blot result shows that pGCSIL-GFP-SMO723 has the highest interfering efficiency (Figure 1).
Figure 1.
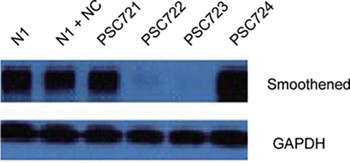
N1 group is 293T cells transfected with pEGFP-N1-SMO; N1 + NC group is 293T cells transfected with pEGFP-N1-SMO and negative control lentivirus plasmid; PSC721 group is 293T cells transfected with pEGFP-N1-SMO and SMO721 recombinant lentivirus plasmid; and PSC722, PSC723, PSC724 groups are 293T cells transfected with pEGFP-N1-SMO and SMO722, SMO723 and SMO724, respectively. GAPHD, Glyceraldehyde-3-phosphate dehydrogenase.
Preparation of SMO RNAi lentivirus particles and titering
At 48 h post co-transfection of the recombinant lentiviral vector pGCSIL-GFP-SMO723 and lentiviral packaging vectors pHelper1.0 and pHelper2.0 in 293T cells, the supernatant was filtered and concentrated. The lentiviral stock was diluted and titered in 293T cells to obtain 2.0 × 108 TU mL−1 (Figure 2).
Figure 2.
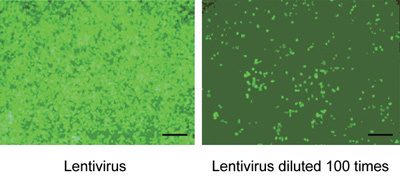
293T cell infected by Lv-SIL-SMO723 after 96 h (the lentivirus diluted at 1:100). Scale bars = 100 μm.
Transfection efficiency assay
At 3 days after the transfection, we used FACSCalibur flow cytometer to measure the efficiency. For the transfection of LNCaP cells, the efficiency of the negative control group was 83.33% and that of the knockdown group was 71.55%. For the transfection of PC3 cells, the efficiency of the negative control group was 83.44% and that of the knockdown group was 88.83% (Figure 3).
Figure 3.
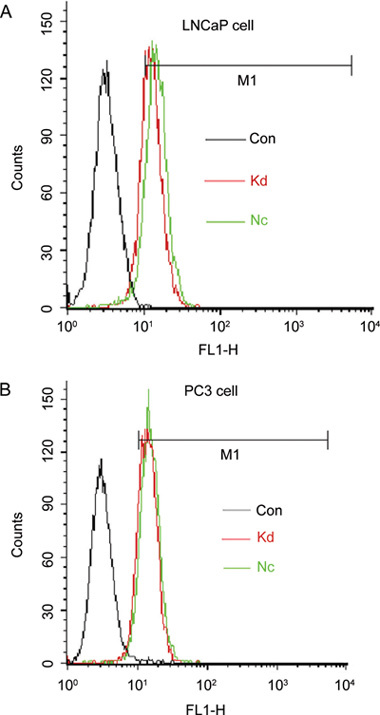
Flow cytometry analysis of cell infection by GFP: LNCaP (A) and PC3 cells (B) were infected with the negative lentivirus or Lv-SIL-SMO723 lentivirus. Con, control group without any infection; Nc, infected with negative lentivirus; Kd, infected with Lv-SIL-SMO723.
qRT-PCR assay for interfering efficiency
Results of the qRT-PCR assay of total RNA show that LNCaP cells in the Lv-SIL-SMO723 knockdown group, compared with the negative control group, have an interfering efficiency of 57.2%; however, PC3 cells in the Lv-SIL-SMO723 knockdown group, compared with the negative control group, have an interfering efficiency of 66.6% (Figure 4).
Figure 4.
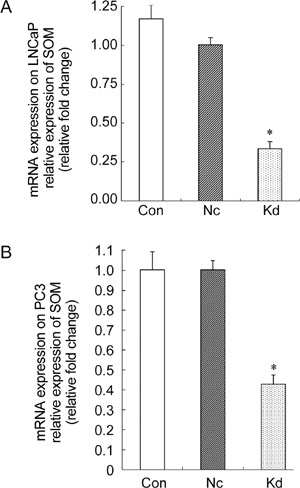
Suppression of smoothened (SMO) mRNA expression by Lv-SIL-SMO723 lentivirus. Cells were infected with the negative lentivirus or Lv-SIL-SMO723 lentivirus, and the RNA expression levels of SMO were detected by quantitative realtime polymerase chain reaction (qRT-PCR). A noticeably decreased SMO mRNA expression (*P < 0.05) was detected compared with the Con group and Nc group, respectively, in both LNCaP (A) and PC3 (B) cell lines. Con, control group without any infection; Nc, infected with negative lentivirus; Kd, infected with Lv-SIL-SMO723.
MTT assay
MTT assay results show that LNCaP cells transfected by Lv-SIL-SMO723 (the knockdown group), when compared with the control group and negative control group, exhibited strong inhibition of cancer cell proliferation (P < 0.01); however, PC3 cells transfected by Lv-SIL-SMO723 did not show a difference among the three PC3 cell groups (Figure 5).
Figure 5.
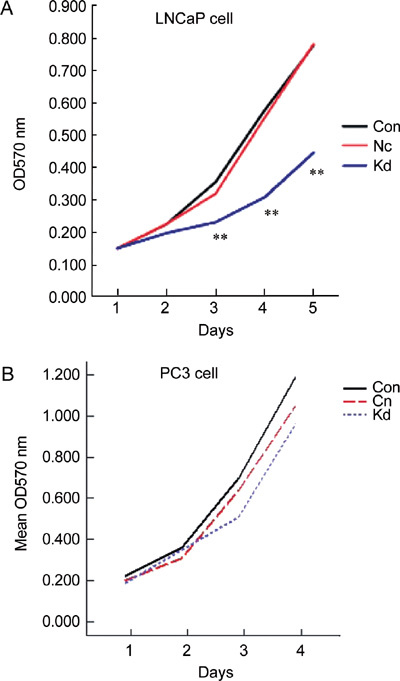
Cell proliferation assay after infection with negative lentivirus and Lv-SIL-SMO723 lentivirus, MTT assay detected the growth of the Con, Nc and Kd groups. (A): A significantly decreased growth rate (**P < 0.01) was detected compared with the Con and Nc groups in the LNCaP cell line. (B): No prominently decreased growth rate (P > 0.05) was detected in the Con and Nc groups in the PC3 cell line. Con, control group without any infection; Nc, infected with negative lentivirus; Kd, infected with Lv-SIL-SMO723.
Cell cycle assay
When compared with the control group and negative control group, cell cycle assay results in both LNCaP and PC3 knockdown groups showed no obvious changes in the cell ratio for the G0/G1 phase, but LNCaP had a higher ratio for the G2/M phase (P < 0.05) and a lower ratio for the S phase (P < 0.05) (Figure 6). Cell divisions of both LNCaP and PC3 cells transfected by Lv-SIL-SMO723 stopped at the G2/M phase.
Figure 6.
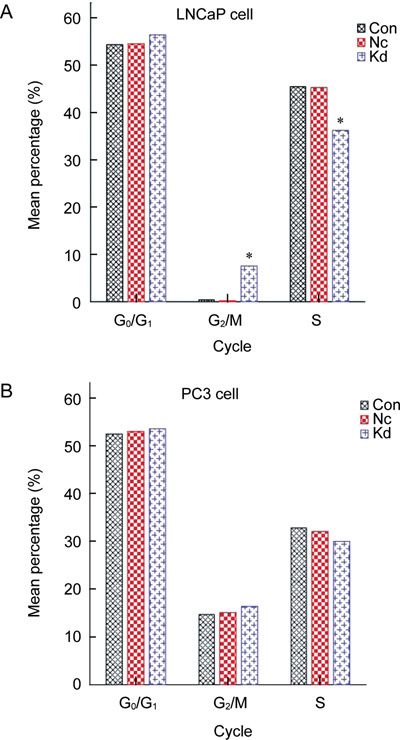
Cell cycles for the Con, Nc and Kd groups were detected by flow cytometry. (A): A pronounced difference was detected compared with the Con and Nc groups in the LNCaP cell line. The mean percentage of S-phase cells was lower (*P < 0.05) and that of G2/M phase cells was higher (*P < 0.05) in the Kd group than in the Con and Nc groups. (B): No prominent difference (P > 0.05) was detected in the Con and Nc groups in the PC3 cell line. Con, control group without any infection; Nc, infected with negative lentivirus; Kd, infected with Lv-SIL-SMO723.
Discussion
Prostate cancer is the leading cancer that endangers men's health. For patients who have no indication of radical prostatectomy, endocrine therapy is the major therapy method. However, many patients have developed androgen-independent prostate cancer 4. A recent study shows that the Hh signaling pathway has an important role in the incidence, growth and metastasis of prostate tumors 5. The Hh signaling pathway, initially discovered by Dr Eric Wieschaus and Dr Christiane Nüsslein-Volhard 6, is a major regulator for cell differentiation and cell proliferation 7. The study by Berman et al. 8 shows that in ventral prostate in mice, the Hh signaling pathway regulates prostate ductal morphogenesis and prostate epithelial differentiation. Many studies show that the growth and survival of prostate cancer rely on the Hh signaling pathway 5, 9. In 2005, the American National Cancer Institute ranked Hh as the first target in prostate cancer therapy, followed by the tumor suppressor gene PTEN and AR 10.
The Hh signaling pathway is composed of Patched (PTCH) and SMO, two transmembrane proteins, and downstream Gli transcription factors. SMO protein, a key transcriber in Hh signaling pathways, can transport Hh signals to Gli signals, and therefore activate nuclear gene transcription and the Hh signaling pathway 11, 12.
Sanchez et al. 9 used Gli-1 RNAi targeting downstream transcription factor Gli-1 in the Hh signaling pathway to inhibit cancer cell development; however, there is no report about RNAi targeting SMO to regulate the activity of Hh signaling pathways to inhibit prostate cancer.
Assuming that SMO can effectively interfere with a target site, this study constructed a recombinant lentiviral vector for RNAi targeting SMO genes, which was then packaged into virus particles and used to transfect the LNCaP and PC3 prostate cancer cell lines, in order to observe its effect on SMO gene expression, cell proliferation and cell cycle.
Results show that the Lv-SIL-SMO723 virus particle can effectively inhibit SMO gene expression in both LNCaP and PC3 cell lines. Infected LNCaP cell proliferation was lower than that of the control and negative control groups (P < 0.05). Infected PC3 cell proliferation showed no difference compared with the control and negative control groups (P > 0.05). The infected LNCaP and PC3 cell division stops at the G2/M phase. Therefore, SMO gene expression regulates prostate cancer cell proliferation and the cell cycle. The lentiviral particles for RNAi, targeting the SMO gene, can specifically suppress SMO gene expression, regulate LNCaP and PC3 cell cycles, and inhibit LNCaP cell growth and proliferation, but has no effect on PC3 cell proliferation. The emergence of the different effects on cell growth between LNCaP and PC3 cells is due to two distinct pathways that are capable of inhibiting cell proliferation. PC3 cell growth can be inhibited mainly through the Cdc25C-Cdc2/cyclin B1 pathway other than the Hh pathway, through which LNCaP cell growth was suppressed 13.
These findings have prompted a novel strategy and targeting site for therapeutic treatment of prostate tumors. In particular, the results that the Lv-SIL-SMO723 virus particle can effectively inhibit SMO gene expression in both LNCaP and PC3 cell lines will be greatly helpful to further research. Furthermore, it would be suggested to use androgen-independent cells to determine simultaneous inhibition of two components of the Hh pathway such as simultaneous Gli-1 inhibition, and how the complicated signaling network comprising the Hh pathway, Wnt pathway and pathway growth factor regulates the carcinogenesis and growth of prostate cancer.
Acknowledgments
This research was funded by National Natural Science Foundation of China (No. 30500506).
References
- Sanchez P, Hernández AM, Stecca B, Kahler AJ, DeGueme AM, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci USA. 2004;101:12561–6. doi: 10.1073/pnas.0404956101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431:707–12. doi: 10.1038/nature02962. [DOI] [PubMed] [Google Scholar]
- Ecke I, Rosenberger A, Obenauer S, Dullin C, Aberger F, et al. Cyclopamine treatment of full-blown Hh/Ptch-associated RMS partially inhibits Hh/Ptch signaling, but not tumor growth. Mol Carcinog. 2008;47:361–72. doi: 10.1002/mc.20394. [DOI] [PubMed] [Google Scholar]
- Shaw G, Prowse DM. Inhibition of androgen-independent prostate cancer cell growth is enhanced by combination therapy targeting Hedgehog and ErbB signalling. Cancer Cell Int. 2008;18:3. doi: 10.1186/1475-2867-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum L, Beachy PA. The hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–9. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Xie J. Hedgehog signaling pathway: development of antagonists for cancer therapy. Curr Oncol Rep. 2008;10:107–13. doi: 10.1007/s11912-008-0018-7. [DOI] [PubMed] [Google Scholar]
- Berman DM, Desai N, Wang X, Karhadkar SS, Reynon M, et al. Roles for Hedgehog signaling in androgen production and prostate ductal morphogenesis. Dev Biol. 2004;267:387–98. doi: 10.1016/j.ydbio.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Sanchez P, Clement V, Ruiz i Altaba A. Therapeutic targeting of the hedgehog GLi pathway in prostate cancer. Cancer Res. 2005;65:2990–2. doi: 10.1158/0008-5472.CAN-05-0439. [DOI] [PubMed] [Google Scholar]
- Vanchieri C. Scientists hopeful as they uncover molecular clues to prostate cancer. J Natl Cancer Inst. 2005;97:168–9. doi: 10.1093/jnci/97.3.168. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumors, embryos and stem cells. Nat Rev Cancer. 2002;2:361–72. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- Murone M, Luoh SM, Stone D, Li W, Gurney A, et al. Gli regulation by the opposing activities of fused and suppressor of fused. Nat Cell Biol. 2000;2:310–2. doi: 10.1038/35010610. [DOI] [PubMed] [Google Scholar]
- Xu XF, Zhang ZY, Ge JP, Cheng W, Zhou SW, et al. RNA interference-mediated silencing of the PAR gene inhibits the growth of PC3 cells via the induction of G2/M cell cycle arrest and apoptosis. J Gene Med. 2007;9:1065–70. doi: 10.1002/jgm.1109. [DOI] [PubMed] [Google Scholar]


