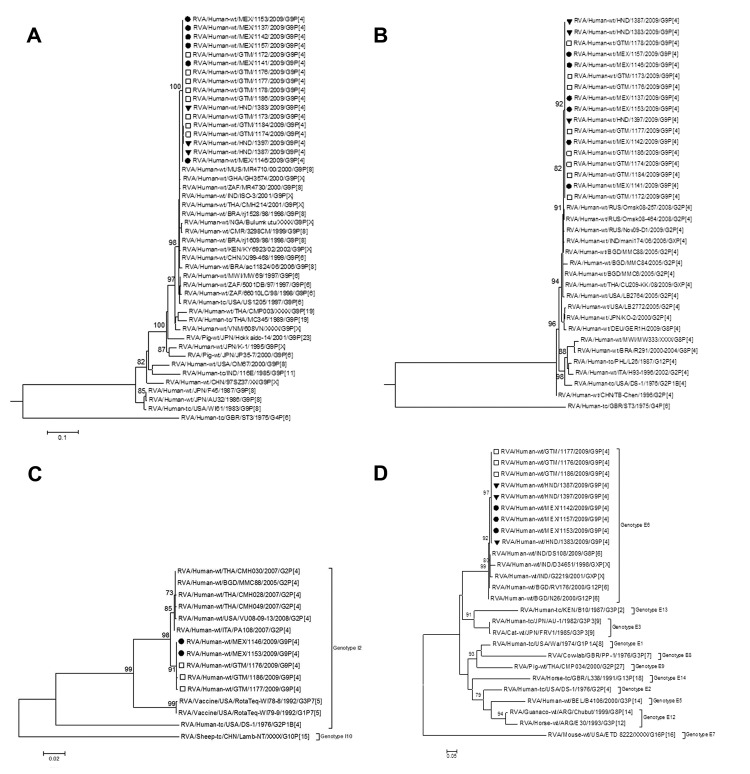
Figure.
Maximum-likelihood phylograms indicating genetic relationships of nucleotide sequences of A) viral protein 7 (VP7), B) VP4, C) VP6, and D) nonstructural protein 4 (NSP4) genes of human G9P[4] group A rotavirus (RVA) strains from Mexico, Guatemala, and Honduras, and sequences of human and animal RVA strains from GenBank. Partial VP4 (VP8* region), VP7 and VP6 gene sequences (742, 783, and 1,155 bases, respectively) and complete gene sequences of NSP4 (528 bases) were aligned with cognate reference strain sequences by using ClustalW in MEGA 5.05 (http://megasoftware.net/mega.php). The optimal evolutionary model that best fit each sequence dataset was identified by using MEGA 5.05. Maximum-likelihood trees were constructed by using SEAVIEW version 4 (www.seaviewfishing.com/DownloadSoftware.html), and approximate likelihood ratio test (aLRT) statistics were computed for estimation of branch support. On the basis of Akaike information criteria with a correction for finite sample sizes, we selected the Tamura-Nei plus gamma, general time reversible plus gamma, general time reversible plus invariant sites, and Hasegawa-Kishino-Yano plus gamma models for genes VP4, VP7, VP6, and NSP4, respectively. Trees are drawn to scale. Only aLRT values ≥70% are shown. Solid circles indicate G9P[4] strains from Mexico, squares indicate G9P[4] strains from Guatemala, and solid inverted triangles indicate G9P[4] strains from Honduras sequenced in this study. Scale bars indicate genetic distances. MEX, Mexico; GTM, Guatemala; HND, Honduras; MUS, Mauritius; GHA, Ghana; ZAF, South Africa; IND, India; THA, Thailand; BRA, Brazil; NGA, Nigeria; CMR, Cameroon; KEN, Kenya; CHN, China; MWI, Malawi; USA, United States; VNM, Vietnam; JPN, Japan; GBR, United Kingdom; RUS, Russia; BGD, Bangladesh; DEU, Germany; PHL, The Philippines; ITA, Italy; BEL, Belgium; ARG, Argentina.