Abstract
The mouse pathobiont Helicobacter hepaticus can induce typhlocolitis in interleukin-10-deficient mice, and H. hepaticus infection of immunodeficient mice is widely used as a model to study the role of pathogens and commensal bacteria in the pathogenesis of inflammatory bowel disease. C57BL/6J Il10−/− mice kept under specific pathogen-free conditions in two different facilities (MHH and MIT), displayed strong differences with respect to their susceptibilities to H. hepaticus-induced intestinal pathology. Mice at MIT developed robust typhlocolitis after infection with H. hepaticus, while mice at MHH developed no significant pathology after infection with the same H. hepaticus strain. We hypothesized that the intestinal microbiota might be responsible for these differences and therefore performed high resolution analysis of the intestinal microbiota composition in uninfected mice from the two facilities by deep sequencing of partial 16S rRNA amplicons. The microbiota composition differed markedly between mice from both facilities. Significant differences were also detected between two groups of MHH mice born in different years. Of the 119 operational taxonomic units (OTUs) that occurred in at least half the cecum or colon samples of at least one mouse group, 24 were only found in MIT mice, and another 13 OTUs could only be found in MHH samples. While most of the MHH-specific OTUs could only be identified to class or family level, the MIT-specific set contained OTUs identified to genus or species level, including the opportunistic pathogen, Bilophila wadsworthia. The susceptibility to H. hepaticus-induced colitis differed considerably between Il10−/− mice originating from the two institutions. This was associated with significant differences in microbiota composition, highlighting the importance of characterizing the intestinal microbiome when studying murine models of IBD.
Introduction
The gram-negative ‘pathobiont’ Helicobacter hepaticus primarily colonizes the gastrointestinal tract of mice, and can also colonize bile canaliculi and gallbladder of mice with chronic hepatitis [1]. While H. hepaticus colonization causes no intestinal pathology in immunocompetent mice, it induces hepatobiliary inflammation and an increased incidence of hepatic cancer in a number of susceptible mouse strains [2]–[4]. In mice with compromised immune function, H. hepaticus in combination with other constituents of the murine intestinal microbiota can induce inflammatory bowel disease (IBD) [5]–[7]. H. hepaticus infections in immunodeficient mice with or without transfer of T cells have been widely used as model systems for human IBD ([5], [8]–[10],for a review see [11]). The H. hepaticus infected Rag −/− mouse model is also widely used as a model of microbially driven colon cancer [12], [13].
Induction of intestinal inflammation in mice depends on a number of known H. hepaticus virulence factors. In addition to traits critical for successful colonization, such as flagellar motility [14], a number of genes and mechanisms directly influencing pathology have been identified. The H. hepaticus genome [15] contains a gene cluster encoding a cytolethal distending toxin (CDT), a homolog of the Campylobacter jejuni CDT, which causes cell cycle arrest, chromatin fragmentation, and apoptosis [16]. It also includes a pathogenicity island HHGI1, which encodes a functional type VI secretion system [17], [18]. Strains carrying the island had a higher potential to induce hepatitis in A/JCr mice [19]. Mutants lacking parts of the HHGI1 island were attenuated with respect to the induction of typhlocolitis in Il10−/− mice [20]. H. hepaticus lipopolysaccharide (LPS) has been shown to reduce Toll-like receptor 4 (TLR4) and TLR5-mediated innate immune responses of intestinal epithelial cells, but also to inhibit development of endotoxin tolerance, thus affecting host responses to the resident microbiota and intestinal inflammatory conditions [21].
Infection with H. hepaticus alone is not sufficient to induce IBD in Il10−/− mice [22], [23]. IBD is induced only in the presence of normal murine microbiota, or during co-infection with specific other bacteria [11], [23]. The effect of the resulting inflammation on the intestinal microbiota depends both on the host immune response and on characteristics of the microbiota, both of which might vary between different mouse strains [7], [24].
An involvement of the intestinal microbiota in the etiology of IBD has been demonstrated in a variety of mouse models. Inflammation induced in response to bacterial infection, a chemical inducer or genetic factors can alter the composition of the intestinal microbiota, which can be accompanied by changes in overall bacterial abundance and diversity [11]. Both in hosts with compromised immune regulation [25] and in mice with deficiencies in colonic epithelial cell inflammasomes [26], these changes can transform the microbiota into a colitogenic state. If this colitogenic microbiota is transmitted to wild type (wt) animals, it can confer a predisposition for inflammation in these healthy hosts [25], [26].
In mouse models of IBD, such as in H. hepaticus infected Il10−/− mice, this dysbiosis and its interdependence with inflammatory processes can be studied in controlled settings. However, even in inbred strains of mice kept under specific pathogen-free (SPF) conditions, the exact composition of the intestinal microbiota is rarely known before the start of the experiment. This study started with the observation that C57BL/6J Il10−/− mice kept at two different animal facilities displayed strongly discordant susceptibilities to H. hepaticus-induced typhlocolitis. We hypothesized that the intestinal microbiota composition might be responsible for these different susceptibilities, and therefore investigated the microbiota compositions of uninfected mice from these two facilities with the aim to identify microbiota components whose differential presence might contribute to or protect the mice from IBD after infection with H. hepaticus. In order to reduce the effect of random differences between groups of mice on our conclusions, we included two groups of MHH mice that were born in different years.
Results
Differential Susceptibility to H. hepaticus-associated Typhlocolitis of C57BL/6J Il10−/− mice Housed in Two Different Animal Facilities
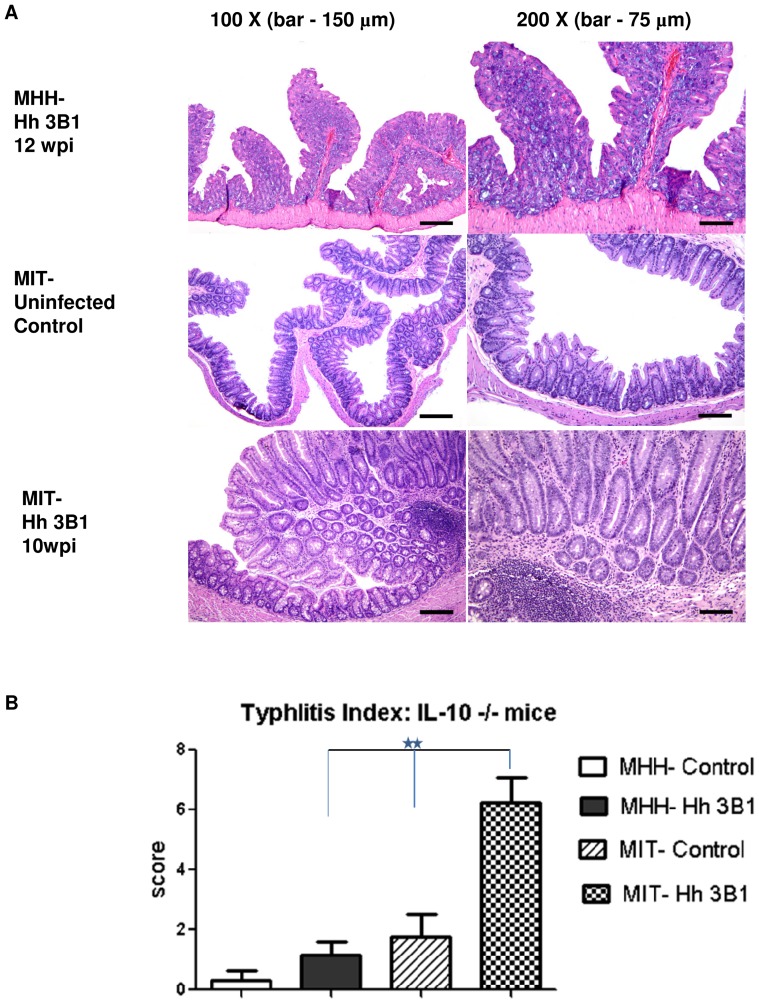
H. hepaticus infected C57BL/6 Il10−/− mice were reported to develop typhlocolitis by the group of J.G.F. at MIT [20], [27], and similar findings have been reported by other groups [6] [28] [29]. By contrast, attempts by the group of S.S. to establish this model at Hannover Medical School (MHH) with the same H. hepaticus strain, ATCC 51449 (3B1), following an identical protocol for inoculation and monitoring of infection, yielded different results. While mice at MIT developed robust typhlocolitis, mice at MHH reproducibly did not develop significant inflammation (Fig. 1), despite a high level colonization with H. hepaticus 3B1 (see Materials and Methods). A potential attenuation of the strain 3B1 due to laboratory passage was ruled out by transfer of a fresh infectious 3B1 isolate from MIT to MHH prior to the experiment depicted in Fig. 1. Furthermore, the strain 3B1 used at MHH caused robust inflammation comparable with results from other laboratories in different (e.g. T cell transfer) models of H. hepaticus-induced colitis [17]. We therefore hypothesized that differences of the intestinal microbiota composition between C57BL/6 Il10−/− mice kept at the two different facilities might be responsible for these observed major differences of susceptibility to H. hepaticus-induced pathology. We therefore performed a high resolution analysis of microbiota composition for C57BL/6J Il10−/− mice from both facilities.
Figure 1. Effects of H. hepaticus infection on MIT vs. MHH mice.
Intestinal inflammation induced by H. hepaticus infection in C57BL/6J Il10−/− mice reared in MIT and MHH SPF facilities. A, histological sections (H&E stain); B, Histological sections of cecum were scored separately for inflammation, epithelial defects, edema, crypt atrophy, hyperplasia and dysplasia on a scale of 0 to 4. The scores from all the different parameters were added and represented as Typhlitis Index scores as described in Burich et al. [29]. P values according to nonparametric Mann-Whitney test, **<0.005.
Microbiota Composition Analysis of C57BL/6J Il10−/− mice Reared at MIT or MHH
We analyzed the intestinal microbiota composition of three groups of Helicobacter-free mice from breeding colonies at MHH and MIT used for H. hepaticus infection experiments, such as the representative experiments described in the previous section. Each group consisted of eight C57BL/6J Il10−/− mice kept under SPF conditions (see Materials and Methods for details). Two of these groups of mice (MHH2009 and MHH2011) were housed at MHH and one group was housed at MIT. Microbiota composition analysis was performed by partial amplification of 16S rRNA genes and deep sequencing using 454 FLX technology (see Material and Methods for details).
We generated a total of 247,219 sequences, which were pre-processed using the mothur pipeline [30]. After a series of quality checks, which included removal of low-quality sequences and of PCR chimeras, 124,930 (51%) sequences were retained. The mean sequence length of the pre-processed sequences was 261 bases. Using the RDP classifier (Bootstrap cutoff 70%) [31], 114,994 of the 124,930 pre-processed sequences (92%) were identified at least to class level, or were assigned to one of the genera unclassified in the RDP dataset (“OD1” and TM7). The remaining sequences were excluded from subsequent analysis following Ochman et al. [32]. No mitochondrial or chloroplast sequences were found in the dataset.
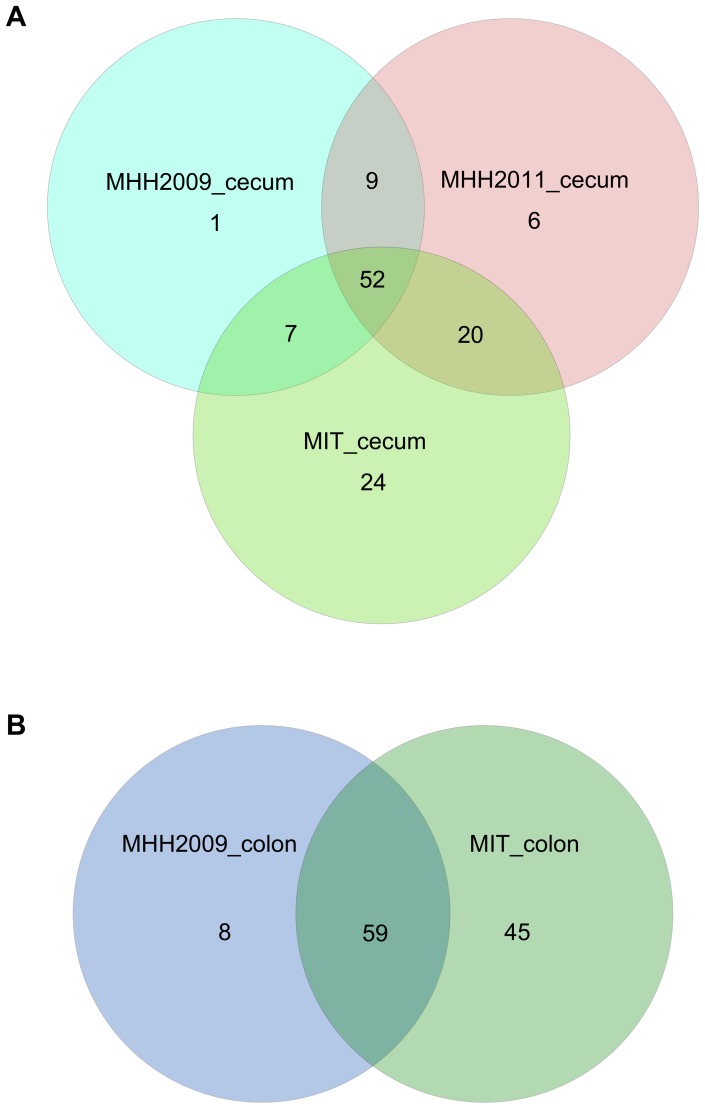
Overall, 28,982 sequences (25% of all retained sequences) were identified to genus at bootstrap support (BTS) 70% (Table S1). Of these, 17,068 sequences (59%) could be further classified to species level using a MegaBlast approach and a modified RDP database. Those sequences were used to choose the distance level of 0.08 as the OTU set for which species and OTU boundaries best correspond (Fig. S1). At this level, the retained 114,994 sequences clustered into 287 OTUs. After subsampling the dataset to 1227 sequences per sample, which was done to ensure equal sequence numbers in all samples, 246 OTUs remained (Table 1 and Table S2). Good’s coverage, which measures the proportion of OTUs sampled at least twice, was 97.5% or higher for all individual samples, and 99.7% or higher for each sample set from one gut region of one group of mice. Of the 246 OTUs retained after subsampling, 119 occurred in at least half the cecum or colon samples of at least one mouse group; the other 127 were considered rare (Table S3). Of the 119 OTUs occurring in at least half the cecum or colon samples of at least one group of mice, 99 (83%) were identified as significantly differently prevalent between all MHH and MIT samples or at least one set of MHH samples and their MIT counterparts (Table S3).
Table 1. OTU counts and Good’s coverage (dataset containing 1227 sequences per sample, 8 samples per sample set).
| Sample set | OTUs | Good’s coverage |
| MIT_cecum | 163 | 99.70% |
| MIT_colon | 173 | 99.70% |
| MHH2009_cecum | 104 | 99.80% |
| MHH2009_colon | 98 | 99.80% |
| MHH2011_cecum | 114 | 99.80% |
| whole dataset | 246 |
Comparisons between Sample Sets
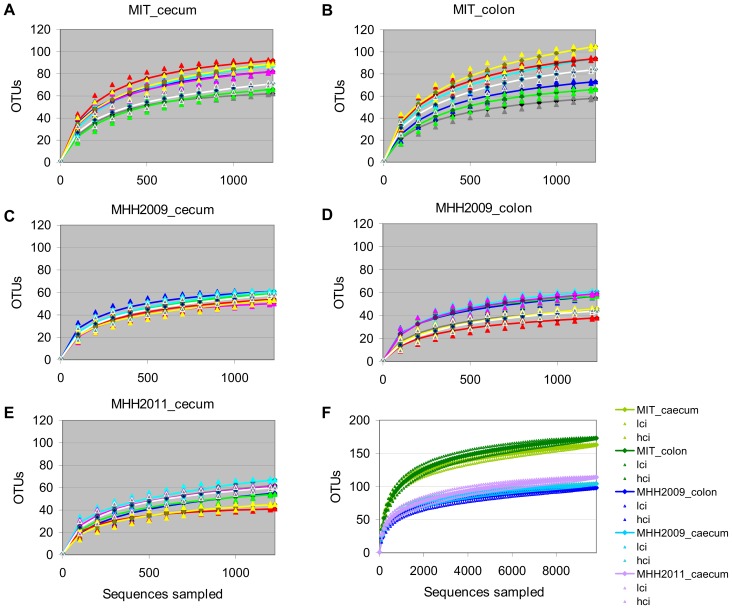
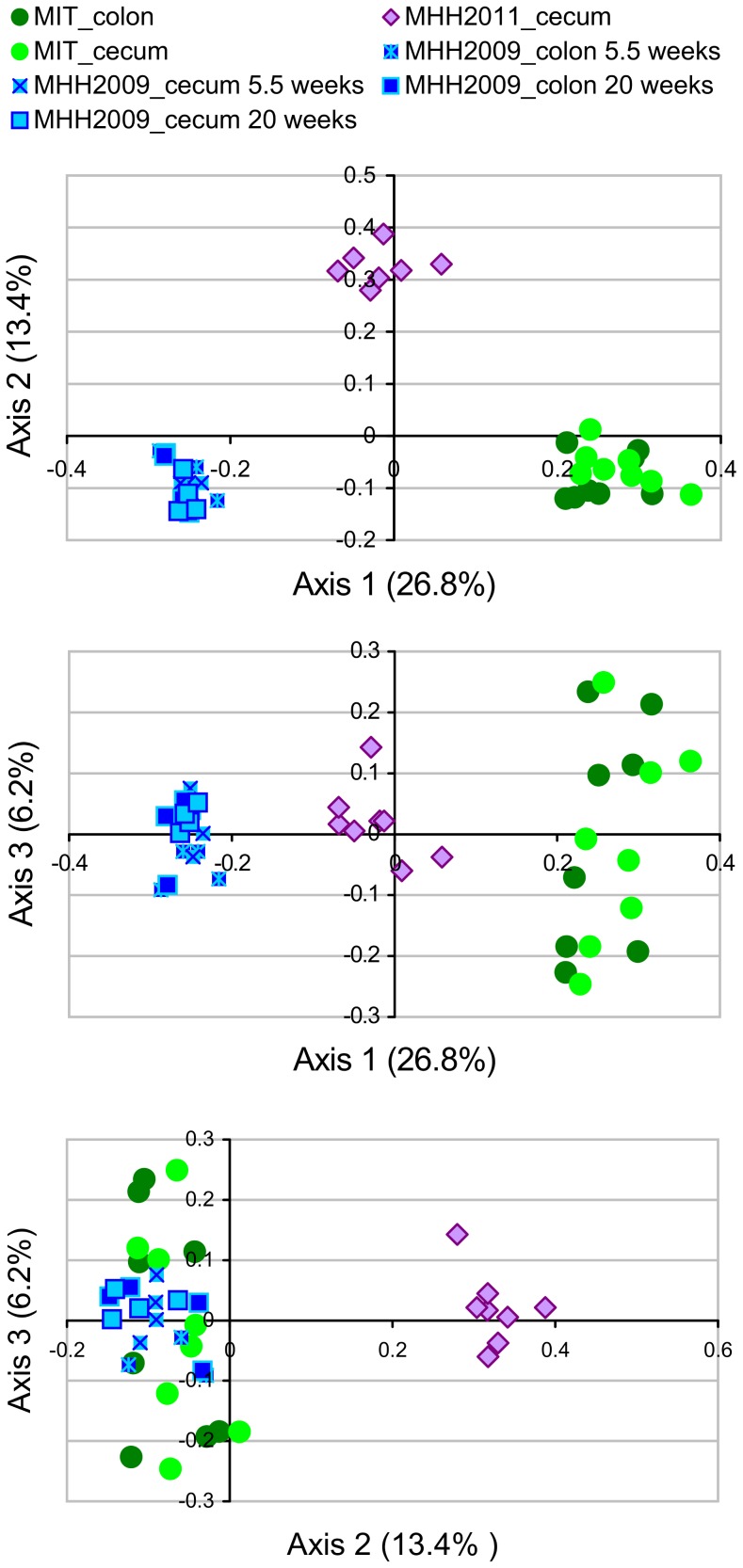
The microbiota richness observed after subsampling to 1227 sequences per sample varied both between mice within one group (Fig. 2A–E) and between mouse groups (Fig. 2F). MIT mice contained considerably more OTUs than MHH animals. Microbiota composition differed between all groups of mice (Fig. 3). In a Principal Coordinates Analysis (PCoA) based on dissimilarity of microbiota compositions, the first two axes, which represented 26.8% and 13.4% of the variance in the dataset, grouped the samples according to mouse group (Fig. 4). While the first axis depicted MHH2011 samples as intermediate between MHH2009 and MIT samples, the second axis emphasized the difference between the MHH2011 and all other mice. The cecum and the colon sample sets of each mouse group were grouped together. Axes 3 to 14, which represented 6.2% to 1.7% of the variance, did not resolve the difference between mouse groups (axes 4 to 14 not shown). Removing the rare OTUs from the dataset did not appreciably change the PCoA results (data not shown).
Figure 2. Rarefaction curves.
A–E, rarefaction curves for individual samples (n = 8 mice), grouped by sample set. F, rarefaction curves for combined data for each sample set (n = 5 sets), including 95% confidence intervals. Lci, lower bound of confidence interval; hci, higher bound of confidence interval. Each sample set consist of either the cecum or the colon samples for one batch of 8 mice. All curves generated after subsampling to 1227 sequences per sample.
Figure 3. Distribution of OTUs occurring in at least 4 samples of the same set.
A, cecum samples, B, colon samples.
Figure 4. Principal coordinates analysis (PCoA) of microbiota samples.
Based on Jaccard index distances measuring dissimilarity of OTU-level microbiota composition between individual samples. Axis labels including fractions of variance explained.
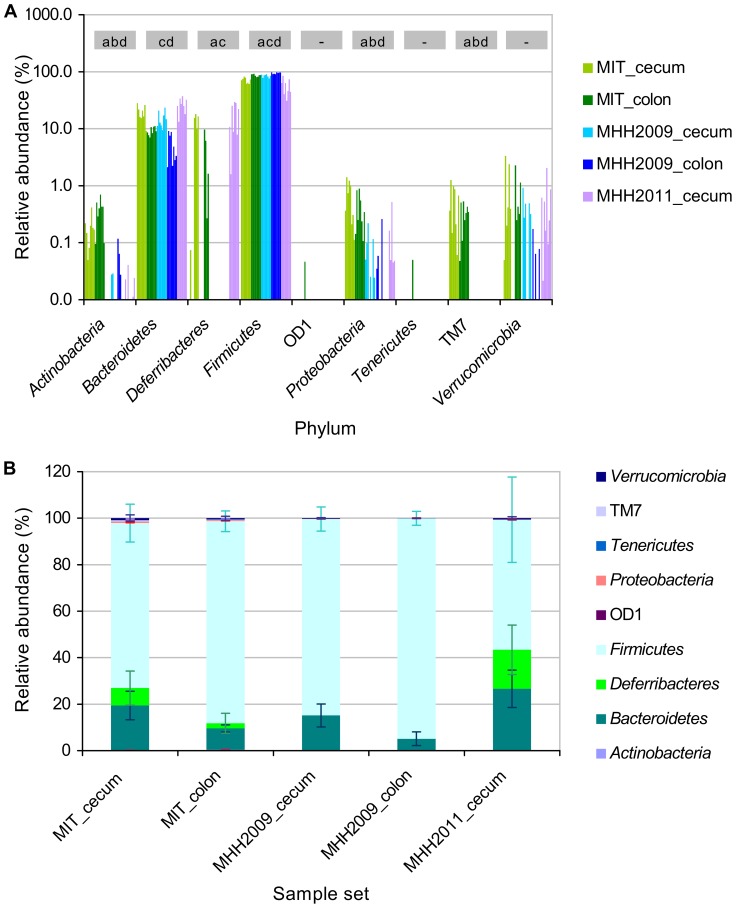
These differences between groups were reflected in the phylum-level composition of the microbiomes (Figure 5 A, B), which was determined by RDP classifier analysis of all 114,994 high-quality sequences. Actinobacteria and Proteobacteria were significantly more abundant in MIT than in MHH samples (two-sided unequal variances t-Test on relative sequence counts, p<0.05). Candidate division TM7, which was represented with only one OTU, was restricted to MIT mice, where it occurred in all samples (see Fig. 5 and Table S3). In MHH2009 samples, Firmicutes accounted for a significantly higher percentage of sequences of both cecum and colon samples than in the two other mouse groups. The difference between MIT and MHH2011 cecum samples was not significant at p≤0.05. Bacteroidetes were significantly less abundant in MHH2009 than in MHH2011 cecum samples, and in MHH2009 colon than in MIT colon samples.
Figure 5. Phylum-level composition of microbiota.
A. Fraction of sequence counts in each of the individual samples, color coded by sample group. Grey boxes indicate significant differences between sample groups (unequal variances t-Test, p<0.05): a, significantly different between MIT and MHH2009 cecum samples; b, significantly different between MIT and MHH2011 cecum samples; c, significantly different between MHH2009 and MHH2011 cecum samples; d, significantly different between MIT and MHH2009 colon samples. B. Relative abundance of phyla in each of the sample groups, including standard deviation (n = 8).
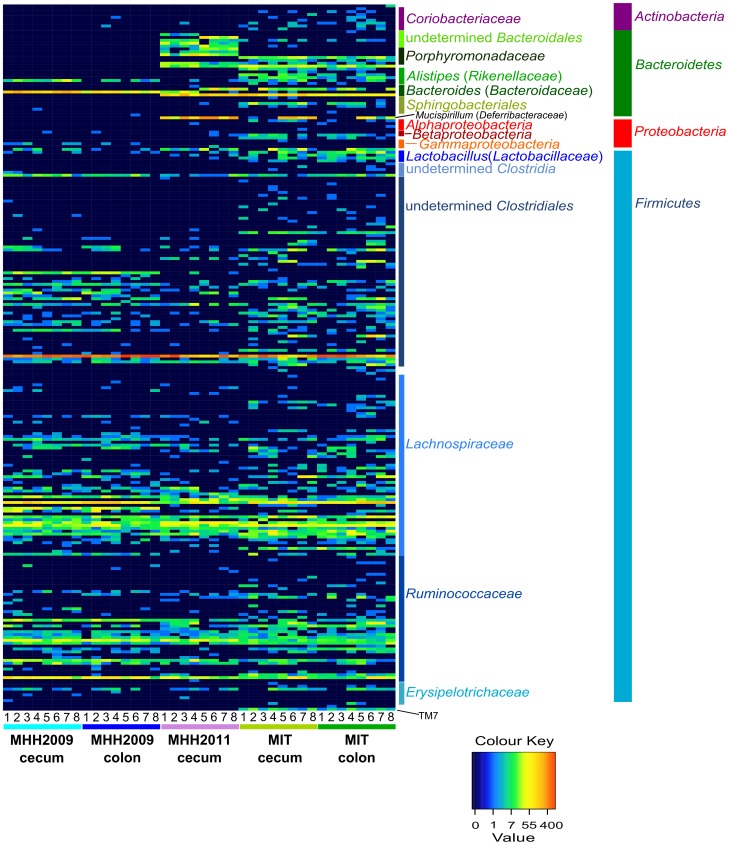
The distribution of Deferribacteres, which were represented by only one OTU classified as Mucispirillum schaedleri, varied considerably among the individual mice of the MIT and MHH2011 groups. They did not occur in MHH2009 mice (Fig. 6 and Table S3).
Figure 6. Distribution of OTUs among samples.
Colors encode absolute OTU counts. Information on OTU classification according to RDP classifier. Numbers 1–8 are mouse identifiers. Within one mouse batch, equal numbers refer to cecum and colon samples of the same animal. Heatmap generated after subsampling to 1227 sequences per sample. The numbers on the color key correspond to untransformed OTU abundances.
Of the 119 OTUs not classified as rare, 24 were only found in MIT samples (Table 2, Table S3). Of these, Parabacteroides merdae, Lactobacillus intestinalis and Clostridium XIVb belonged to genera also identified from the sequences of the altered Schaedler flora (ASF) as provided by Dewhirst et al. [33] (Table S4). The most abundant MIT-specific OTU was classified as Odoribacter, a member of the Porphyromonadaceae within the Bacteroidales. 13 of the 24 OTUs were members of the Clostridiales. Eight of these could be further classified as Lachnospiraceae, including the Clostridium XIVb OTU mentioned above. Four of the MIT-specific OTUs were members of the Rikenellaceae genus Alistipes. This genus was also represented by a rare OTU only found in MIT samples (but not included in the list of MIT-specific OTUs because it was only found in 3 mice) and a further OTU classified as Alistipes finegoldii that occurred in all mouse groups Other MIT-specific OTUs were classified as Bilophila wadsworthia (Desulfovibrionaceae), a member of the Sphingobacteriales, and a member of the candidate division TM7.
Table 2. MIT-specific OTUs occurring in at least 4 samples of one MIT sample set.
| OTU label | OTU classification | Number of reads assigned to OTU(samples with OTU) | |
| MIT, cecum | MIT, colon | ||
| 0.08_group_254 | Bacteroidetes, Bacteroidia, Bacteroidales, Porphyromonadaceae, Odoribacter | 141 (8) | 104 (8) |
| 0.08_group_249 | Firmicutes, Clostridia, Clostridiales | 49 (3) | 172 (4) |
| 0.08_group_243 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae, Clostridium XlVb | 61 (7) | 69 (8) |
| 0.08_group_126 | Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes | 71 (7) | 37 (7) |
| 0.08_group_106 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 60 (7) | 38 (7) |
| 0.08_group_191 | TM7, genera, incertae sedis | 58 (8) | 38 (8) |
| 0.08_group_120 | Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes | 72 (6) | 21 (5) |
| 0.08_group_286 | Firmicutes, Clostridia, Clostridiales | 37 (4) | 52 (3) |
| 0.08_group_270 | Firmicutes, Clostridia, Clostridiales | 37 (7) | 25 (5) |
| 0.08_group_104 | Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes | 49 (8) | 11 (5) |
| 0.08_group_195 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 8 (4) | 33 (6) |
| 0.08_group_141 | Bacteroidetes, Bacteroidia, Bacteroidales, Porphyromonadaceae, Parabacteroides, Parabacteroides merdae | 22 (7) | 15 (5) |
| 0.08_group_131 | Proteobacteria, Deltaproteobacteria, Desulfovibrionales, Desulfovibrionaceae,Bilophila, Bilophila wadsworthia | 20 (6) | 10 (4) |
| 0.08_group_221 | Firmicutes, Clostridia, Clostridiales | 13 (6) | 15 (5) |
| 0.08_group_107 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 23 (6) | 5 (2) |
| 0.08_group_188 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 7 (3) | 20 (4) |
| 0.08_group_246 | Firmicutes, Bacilli, Lactobacillales, Lactobacillaceae, Lactobacillus, Lactobacillus intestinalis | 5 (3) | 16 (5) |
| 0.08_group_153 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 11 (4) | 9 (4) |
| 0.08_group_156 | Firmicutes, Clostridia, Clostridiales | 7 (4) | 9 (5) |
| 0.08_group_97 | Proteobacteria, Alphaproteobacteria | 13 (5) | 3 (3) |
| 0.08_group_119 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 9 (3) | 7 (4) |
| 0.08_group_144 | Bacteroidetes, Sphingobacteria, Sphingobacteriales | 6 (4) | 6 (4) |
| 0.08_group_118 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 2 (1) | 9 (5) |
| 0.08_group_260 | Bacteroidetes, Bacteroidia, Bacteroidales, Rikenellaceae, Alistipes | 9 (4) | 1 (1) |
No conclusive pattern could be detected among the MHH-specific OTUs (Table 3), as all OTUs not occurring in MIT mice were either non-existent in one of the MHH groups, or occurred in less than half of the samples and were thus classified as rare in either MHH2009 or MHH2011 mice (Fig. 6, see Table S3).
Table 3. MHH-specific OTUs occurring in at least 4 samples of one MHH sample set.
| OTU label | OTU classification | Number of reads assigned to OTU (samples with OTU) | ||
| MHH2009, cecum | MHH2009, colon | MHH2011, cecum | ||
| 0.08_group_252 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 224 (8) | 222 (8) | 5 (2) |
| 0.08_group_169 | Firmicutes, Clostridia, Clostridiales | 116 (8) | 127 (8) | 7 (2) |
| 0.08_group_199 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 64 (4) | 50 (4) | 36 (3) |
| 0.08_group_130 | Firmicutes, Clostridia, Clostridiales | 24 (4) | 24 (4) | 6 (3) |
| 0.08_group_121 | Firmicutes, Clostridia, Clostridiales, Ruminococcaceae | 13 (5) | 9 (4) | 2 (2) |
| 0.08_group_117 | Proteobacteria, Gammaproteobacteria, Pseudomonadales, Moraxellaceae, Acinetobacter | 10 (5) | 3 (2) | 0 (0) |
| 0.08_group_234 | Firmicutes, Clostridia, Clostridiales, Lachnospiraceae | 1 (1) | 1 (1) | 35 (7) |
| 0.08_group_155 | Bacteroidetes, Bacteroidia, Bacteroidales | 0 (0) | 0 (0) | 18 (4) |
| 0.08_group_170 | Bacteroidetes, Bacteroidia, Bacteroidales, Porphyromonadaceae | 0 (0) | 0 (0) | 36 (7) |
| 0.08_group_157 | Bacteroidetes, Bacteroidia, Bacteroidales, Porphyromonadaceae | 0 (0) | 0 (0) | 79 (6) |
| 0.08_group_198 | Bacteroidetes, Bacteroidia, Bacteroidales | 0 (0) | 0 (0) | 123 (7) |
| 0.08_group_168 | Bacteroidetes, Bacteroidia, Bacteroidales | 0 (0) | 0 (0) | 244 (4) |
| 0.08_group_184 | Bacteroidetes, Bacteroidia, Bacteroidales, Porphyromonadaceae | 0 (0) | 0 (0) | 317 (8) |
Discussion
H. hepaticus-infected immunodeficient mice are a widely used model system for the investigation of inflammatory bowel diseases [12]. While monoassociation with H. hepaticus has been reported to cause sporadic colitis in mono-associated Swiss Webster mice during very long-term infection [3], that does not normally hold true for the widely used Il10−/− mice [7], [22]. Experiments using the Il10−/− H. hepaticus-induced colitis model are usually conducted in the presence of a normal mouse microbiota or SPF microbiota [20], [28], [29]; the exact composition of which remains mostly unknown. In spite of the widespread use of this model, different outcomes of H. hepaticus infection in Il10−/− mice are well known [6], [22]. Here we compared the microbiota of C57BL/6 Il10 −/− mice that were reared at two different institutions and showed highly reproducible differences in their susceptibility to H. hepaticus-induced pathology.
We found major differences between the microbiota of the mice housed in the two different animal facilities. There was also significant (but much lower) microbiota variation between the two groups of MHH mice (Fig. 4), which had originated from the same breeding colony but were born in different years. Multiple factors may have contributed to the differences between the microbiota of the three mouse groups. These include differences in animal husbandry (e.g. chow, cohousing with other mouse groups) and the propagation of random effects during the seeding and development of the individual microbiota. In a study mainly searching for genetic factors influencing microbiota in mice, Benson et al. [34] also found significant effects of the litter of origin, even when accounting for the identity of the mother. Hufeldt et al. [35] detected significant differences between the cecal microbiota of mouse litters by dams of the same breeding colony, but significantly less variation if these dams were siblings from the same litter. Finally, minor genetic differences between the two colonies of mice which have been bred for several years at different facilities cannot be excluded and may also contribute to differences of microbiota composition.
Age has been reported as a determinant of microbiota composition in mice [36], [37]. The MHH2009 group contained mice of two different ages. An AMOVA analysis performed with mothur 1.27.0 found significant differences of microbiota composition between two subgroups of the MHH2009 groups that were aged 1 vs. 4 months (data not shown). However, this age effect was minor compared to the differences between MHH-raised and MIT-raised mice. The PCoA clustering of the same data confirmed that the effects of age were relatively minor (Fig. 4) while the differences between MHH and MIT-reared mice were substantial. This is consistent with the observation that the absence of H. hepaticus-induced pathology was reproducibly observed at the MHH facility when C57BL/6 Il10−/− mice of different ages (10–17 weeks at the time of first infection) were infected with H. hepaticus 3B1 (data not shown).
In the current study, colon and cecum samples clustered together for both MIT and MHH2009 mice (Fig. 4). This effect is also known from humans, where the microbiota between cecal and colonic mucosal samples from the same study participant tend to be rather similar [38], while considerable differences exist between individuals [38], [39].
In order to identify intestinal bacterial candidates that might either contribute to colitis development in MIT mice or protect the MHH mice from intestinal inflammation [40], [41], we compared the intestinal microbiota of both groups of MHH mice to their counterparts from the MIT.
Surprisingly, all MHH-specific (approximately species-level) OTUs were either not detected or classified as rare in at least one of the sample sets. We could however identify five OTUs that occurred in all groups of mice and that were significantly more abundant in MHH mice than in MIT mice (Table S3). In addition to the three Lachnospiraceae OTUs classified only to family level; the OTUs included Bacteroides intestinalis and an OTU of the genus Oscillibacter, which are both constituents of a healthy gut microbiota in humans [42], [43]. Notably, another Oscillibacter OTU was significantly more abundant in the corresponding MIT samples.
Of the group of 24 OTUs found to be MIT-specific, several were classified as taxa that are associated with healthy microbiota in humans, such as Odoribacter, Parabacteroides merdae, and Alistipes [42]–[45]. One OTU was identified as Clostridium XlVb, a taxon also identified from altered Schaedler flora (ASF) sequences (Table S4), implying that the corresponding organism is closely related to strain ASF 356 in the widely used defined 8-strain rodent ASF [33].
MIT-specific OTUs also included the one OTU of candidate division TM7 found in our dataset. Certain phylotypes within this lineage are associated with IBD in humans [46], and TM7 were also abundant in a transmissible colitogenic microbiota in inflammasome-deficient mice [26]. The two other most prominent components of that colitogenic community were two phylotypes of the family Prevotellaceae [26]. The only OTU of this family in the present dataset was lacking in all MHH2009 samples and occurred significantly more abundantly in the MIT caeca compared to those of the MHH2011 mice (Table S3).
Another noteworthy MIT-specific OTU was the one identified as Bilophila wadsworthia. While this species occurs in the intestinal microbiota of healthy humans [47], it can also act as an opportunistic intestinal pathogen, for example the organism is present in human cases of appendicitis [48]–[50]. Intestinal blooms of B. wadsworthia, which can be induced by dietary interventions (saturated fatty acid-rich diets such as enhanced in milk- or other animal-derived fat), can lead to significantly increased colitis in both mono-associated and SPF C57BL Il10−/− mice [51]. This might also be considered as one factor influencing microbiota between the MIT and MHH facilities, since MIT feed contained 5% porcine fat, while the MHH feed used in both the breeding and experimental facilities was virtually free from animal fat.
An additional microbial component that should be mentioned in the context of this model is Lactobacillus reuteri, which occurred with one OTU in our dataset. It was extremely rare in MHH mice – we identified a single sequence across all MHH samples – but was present in all MIT mice. Lactobacillus species are known to induce the production of both pro- and anti-inflammatory cytokines [52], and in vitro as well as in vivo effects tend to vary between strains [53]. In different experiments, Lactobacillus cocktails containing L. reuteri have been found to both stimulate immune response in piglets [54] and to reduce H. hepaticus-associated inflammation in Il10-deficient mice [40]. If newly introduced into Lactobacillus-free wt mice, L. reuteri alone can trigger non-pathological low-level inflammatory processes in the small intestine [55]. As shown by Whary et al. [23], co-colonization with L. reuteri alone can be sufficient to trigger the development of typhlocolitis in B6-Il10−/− mice infected with H. hepaticus.
Given these components of the MIT microbiota, IBD in MIT mice might be initiated by the combined action of H. hepaticus and the reasonably abundant L. reuteri. The initial inflammatory response in the lower bowel might be intensified by a bloom of the opportunistic pathogen Bilophila wadsworthia and potentially further exacerbated by a combined expansion of the Prevotellaceae and TM7 OTUs; the combination of these bacteria leading to a colitogenic state of the microbiota.
This study of the intestinal microbiota composition of mice with differential susceptibility to H. hepaticus-induced colitis permits the generation of a testable hypotheses; which of the microbial components that might be responsible for increasing inflammation in MIT mice or dampening inflammation in MHH mice? For example, future studies should verify the roles of both L. reuteri and B. wadsworthia in colitis development, and investigate whether the presence of one or both of these species in non-rare numbers in combination with the MHH microbiota would lead to colitis development during H. hepaticus infection.
Materials and Methods
Mice
We analyzed the intestinal microbiota composition of three groups of mice that were not infected with H. hepaticus. Each group consisted of 8 C57BL/6J Il10−/− (B6.129P2-Il10tm1Cgn/J or B6.129P2-Il10tm1Cg/JZtm, respectively) mice kept under SPF conditions. Two of these groups (MHH2009 and MHH2011) were housed at Hannover Medical School (MHH), and one group was housed at Massachusetts Institute of Technology (MIT). MHH animals were derived from a nucleus colony regularly screened during routine genetic monitoring [56]. The MHH2009 group consisted of mice sacrificed at 38 (4 mice) or 141 (4 mice) days of age (MHH2009). MIT mice were necropsied at 172 days of age, and MHH2011 mice at 204 days of age. MHH2009 and MHH2011 mice were from the same breeding colony used as a source of mice for H. hepaticus infection experiments, both groups were necropsied approximately 16 months apart. From the colocecal junction of each MHH2009 and MIT animal, we obtained one colon and one cecum sample. From MHH2011 animals, only cecum samples were included. The samples were snap-frozen in liquid nitrogen before being placed at −80°C.
H. hepaticus Infections
H. hepaticus infections with the wild type strain ATCC 51449 (3B1) were performed and monitored exactly as described previously [17]. Infections at MHH and MIT were performed with the same H. hepaticus strain, and a H. hepaticus strain exchange between MIT and MHH was performed to rule out that differences between strains were the reason for the observed differences in pathology. C57BL/6J Il10−/− mice were bred at the animal facility of Hannover Medical School under Helicobacter-free SPF conditions. Access to mice was limited to few animal caretakers who had to pass through a water shower and were required to wear a gown, cap, surgical mask, overshoes, and gloves. Mice were housed separated by sex in open cages (540 cm2 floor area) at a maximum of five animals on bedding of autoclaved, dust-free, softwood fibers. Sterile pelleted diet (ssniff R-Z, ingredients include vegetable protein and fat, but no animal-derived ingredients) and deionized, filtered, and UV light-treated water were provided ad libitum. The breeding colony was regularly monitored for the presence of common murine pathogens and various Helicobacter species according to FELASA recommendations [57], and all animals were tested again for the presence of Helicobacter species before the start of an experiment. For infection experiments, animals were transferred to an S2 infection unit, where animals were kept in ventilated isolator cages under SPF conditions and fed sterile water and irradiated chow (ssniff M-Z feed; ingredients include soy protein and vegetable fat, but no animal-derived ingredients) ad libitum. All experiments at MHH involving mice were conducted in accordance with the German animal protection law and with the European Communities Council Directives 86/609/EEC and 2010/63/EU for the protection of animals used for experimental purposes. All experiments at MHH were approved by the MHH Animal Protection Officer (“Tierschutzbeauftragter der MHH”) and permitted by the local authority (Lower Saxony State Office for Consumer Protection, Food Safety, and Animal Welfare Service, AZ 33.9-42502-04-08/1456). At MIT, Il10 −/− C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) were maintained free of known murine viruses, Salmonella spp., Citrobacter rodentium, ecto- and endoparasites, and known Helicobacter spp. in a facility accredited by the Association for Assessment and Accreditation of Laboratory Care International, under barrier conditions. All experiments performed at MIT were approved by the “MIT Committee on Animal Care”. Animals were housed in microisolator, solid-bottomed polycarbonate cages on hardwood bedding, fed a commercial pelleted diet (Prolab RMH 3000; ingredients include fish meal and 5% porcine fat conserved with BHA), and administered water ad libitum. The protocol was approved by the MIT Committee on Animal Care.
To monitor the infection status throughout the experiment, fecal pellets were collected once a week and the presence or absence of H. hepaticus was assessed by semi quantitative PCR from total DNA isolated from the stools (Tissue Amp Kit, Qiagen Inc.). Usually, no weight loss or any other external signs of disease or ill health upon visual inspection of the animals were observed throughout the observation period. Occasionally, loose stools were observed with the infected animals.
After necropsy, tissue samples from both cecum and colon were weighed, homogenized and plated on selective blood plates for H. hepaticus cfu counts per mg tissue (MHH), or DNA was extracted for quantitative PCR (MIT) [58], [59]. CFU counts from MHH infection experiments and qPCR-derived H. hepaticus genome counts from the corresponding MIT infections yielded similar results. At MHH, H. hepaticus 3B1 infection of mice from the same breeding colony and born within 6 days of the MHH2011 animals resulted in 5.5 * 107 to 6.1 * 108 cfu per g cecum. At MIT, the qPCR analysis of cecal samples from 3B1-infected mice yielded 103 to 106 genome copies per µg mouse DNA, corresponding to 105–108 bacteria per g tissue [20]. Samples from the cecum and colon were also fixed and embedded for tissue sectioning and pathological scoring.
Mouse Pathology
Tissue sections were scored according to the scoring system developed by Burich et al. [29] by blinded pathologists who are experts in rodent intestinal pathology. To obtain the direct comparison shown in Fig. 1, unstained slides of samples from both facilities were exchanged and read by one pathologist who was blind to the sample identity.
DNA Extraction
DNA was extracted using the QIAamp DNA Mini Kit (Qiagen). Samples were lysed overnight in Buffer ATL supplemented with Proteinase K (both Qiagen) at 56°C, and DNA was extracted according to the QIAamp DNA Mini Kit tissue protocol. DNA was quantified on a NanoDrop 1000 spectrophotometer (Thermo Scientific).
Partial Amplification and Deep Sequencing of 16S rRNA Genes
For MHH2009 and MIT samples, 16S V2 amplicons for 454 pyrosequencing were prepared as described by Turnbaugh et al. [60]. 16S ribosomal DNA gene fragments were amplified using 454 FLX fusion primers containing 454 adapter sequences, linker nucleotides, multiplex identifiers (MIDs) and template-specific parts consisting of the universal bacterial primers 8F (5′- AGAGTTTGATCCTGGCTCAG-3′) and 338R (5′-TGCTGCCTCCCGTAGGAGT-3′). PCR reactions contained 30–50 ng DNA, 0.3 µM of each primer, 2.5 mM Mg2+, 0.2 mM of each dNTP, and 2.5 U HotMaster Taq Polymerase (5PRIME) per 50 µl reaction. Cycling conditions were 95°C for 2 min followed by 30 cycles of 20 s at 95°C, 20 s at 52°C, and 1 min at 65°C. PCRs included a positive control and a no-template negative control, and PCR success was confirmed on 1% agarose gels. Amplicons were purified with AMPure beads (Agencourt, Beckman Coulter) as described in the 454 FLX User Manual and quantified with a NanoDrop spectrophotometer. Length and amplicon integrity were controlled with Agilent DNA 1000 Chips on a 2100 Expert Bioanalyzer (Agilent). Sets of 8 samples with different MIDs were pooled at 200 ng per sample. Emulsion PCR was carried out and samples were sequenced from the 3′ end according to the 454 FLX manual. Raw data were processed with the amplicon pipeline of GS Run Processing Software version 2.3 (454 Life Sciences) using default settings.
For the MHH2011 cecum samples, 16S ribosomal DNA gene fragments were amplified as described in Lofgren et al. [61] 454 FLX Titanium fusion primers included MIDs and template-specific parts consisting of the universal bacterial primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 541R (5′-WTTACCGCGGCTGCTGG-3′). The primer pair was tested for compatibility with primer pair 8F-338R using the SILVA database tool TestPrime (v. 1.0, database SSU r114 RefNR) [62] in combination with custom vba and sql code (Table S5). The broad agreement between the sets of sequences matching each primer pair indicated that the differences between the microbiota of MHH2011 mice on the one hand and MIT and MHH2009 mice on the other hand are unlikely to be due to primer differences. PCR reactions contained 30 ng DNA, 0.3 µM of each primer, 2.5 mM Mg2+, 0.2 mM of each dNTP, and 2.5 U HotMaster Taq Polymerase (5PRIME) per 50 µl reaction. Cycling conditions were adapted following the 454 FLX Titanium manual: 95°C for 2 min followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 80 s at 72°C. PCRs included a positive control and a no-template negative control. PCR products were run on 1% agarose gels, and 650–700 bp fragments were extracted using the QIAquick Gel extraction kit (Qiagen). Extracted amplicons were quantified using the Quant-iT PicoGreen dsDNA Kit (Invitrogen) on a TBS-380 Fluorometer (Turner BioSystems). Sets of 8 samples with different MIDs were pooled at 40 ng per sample. Emulsion PCR was carried out using the emPCR Titanium Lib-L kit. Samples were sequenced from the 3′ end using the Titanium Sequencing Kit XLR70 according to the 454 FLX Titanium manual (version of 20th October 2009). Raw data were processed with the amplicon pipeline of GS Run Processing Software version 2.3 as above, but with the TrimBackScaleFactor set to 2∶160. Sequences were submitted to the European Nucleotide Archive (ENA) under project accession number PRJEB4164 (secondary study accession number ERP003425).
Sequence processing and bioinformatic analysis
Sequences were pre-processed using the software package mothur [30] (Linux 32-bit version 1.22.1 and Windows 32-bit version 1.21.1) following the suggestions in the “Schloss SOP” tutorial [63], [64]. Briefly, primers and barcodes were removed, and sequences were trimmed where the average quality score over a 50-bp-window dropped below 35. Sequences were considered low quality and culled if they contained homopolymer stretches longer than 8 bp, included ambiguous base calls, or were shorter than 150 bases. Remaining non-identical sequences were aligned to the “SILVA” reference alignment [65] with mothur’s align.seqs algorithm, using the default settings of 8-mer searching and Needleman-Wunsch alignment. Sequences which did not align in the expected region of the reference alignment were removed. PCR chimeras were identified using UCHIME [66] with the mothur-provided “SILVA gold” reference set [65], and culled.
The resulting dataset was analyzed with Ribosomal Database Project (RDP) classifier version 2.4 with the hierarchy model of training data no. 7 [31]. Sequences were classified to the lowest taxonomic rank that received at least 70% bootstrap support (BTS). Following Ochman et al. [32], sequences which could not be assigned to class at BTS 70% were excluded from further analyses, except when they were classified to lower taxonomic ranks for which no class was designated. Potential organellar sequences were further analyzed by blastn similarity search against the SILVA reference database, build 108 [67]. In order to obtain the possibility to reliably identify sequences to species, the RDP 16S rRNA database (release 10, update 28, matching RDP classifier training data no. 7) was modified using TaxCollector [68] to include taxonomic descriptions with easily identifiable species designations. Query sequences identified to genus level by RDP classifier at a stringent cutoff of 97% BTS were searched against this TaxCollector-modified RDP database using MegaBlast [69]. Sequences were considered as annotated to species level if their identity to the best hit in the database was at least 97%, the aligned region covered at least 97% of the query sequence length, they did not match a second species at the same BLAST Expectation value, and the identified species did not conflict with the genus as identified by RDP classifier.
In order to facilitate comparison of these data with the altered Schaedler flora, the sequences provided by Dewhirst et al. [33] were similarly analyzed by RDP classifier.
OTUs were calculated using ESPRIT-Tree [70]. In order to avoid artifacts due to the inclusion of the additional sequence stretch in the longer MHH2011 sequences, all sequences were first trimmed to the region corresponding to the 8F-338R amplicon as identified in the mothur-generated alignment. Following Cai and Sun [70], the cutoff OTU level to best represent species level was determined using the normalized mutual information (NMI) criterion [71] on a subset of sequences that could be identified to species. For sequences classified to species, the NMI between species and OTU classification was calculated following Fred and Jain [71]. After comparing NMI values for OTUs obtained for 30 distance levels from 0.01 to 0.3 (Fig. S1), we retained the OTUs at level 0.08, which resulted in peak NMI values. To link all sequences in the dataset with both OTU and taxonomic information, OTUs were classified to the lowest possible rank based on the 80% consensus of their respective RDP classifications using a custom VBA script. Additionally, OTU consensus sequences were computed in Geneious Basic v. 5.6.4 [72] based on alignments generated using MUSCLE [73]. For alignment of OTU 0.08_group_279, which contained too many sequences to be computed in MUSCLE, we used MAFFT [74] with the settings “FFT-NS-2–ep 0.123”. Consensus sequences were classified using the RDP classifier and the TaxCollector-modified RDP database as detailed above. As OTU consensus sequences could often be classified to a lower taxonomic rank than the majority of their component sequences, but were only crudely classified if they contained ambiguous bases, we chose the more detailed of the two classifications for each OTU (Table S6).
To further analyze and visualize the final dataset, a mothur-compatible “sharedfile” was reconstructed from the OTU information. The Windows version of mothur (version 1.21.1) was used for a range of statistical and visualization steps: To avoid artifacts produced by unequal sample sizes, the sequences from each mouse sample were subsampled to match the 1227 sequences found in the sample with the lowest sequence count. Rarefaction curves and rank-abundance-plots were generated. Differences among samples were visualized by Principal Coordinates Analysis (PCoA) using Jaccard index distances, which measure dissimilarity of microbiome composition. Mothur was also used to generate Venn diagrams (see below).
A heatmap of OTU abundances among samples was produced using the R script heatmap [75]. Input data for heatmap construction was based on the mothur-type sharedfile, which was transformed to facilitate visualization of the full range of OTU abundances. This transformation was carried out according to the formula h = LN(count+0.1), where h is the input value for the heatmap script, count is the sequence count for one OTU in an individual sample, LN is natural logarithm and 0.1 is used as a small increment to avoid occurrences of LN(0). The heatmap color key was manually modified to correspond to untransformed OTU abundances. To increase readability, individual OTU labels were replaced with taxonomic information and sample labels were replaced with sample numbers and group information using Inkscape 0.48.
Sequences which did not occur in at least 4 samples of either the cecum or the colon samples of at least one mouse group were classified as too rare to judge site specificity and excluded from the lists of site-specific OTUs and from the Venn diagrams. For additional comparisons, significance of abundance differences between sample groups was assessed using Metastats [76], as implemented in the original R script, at a significance cutoff of p≤0.05.
Supporting Information
Normalized mutual information (NMI) value between species and OTUs obtained for 30 difference levels using ESPRIT-Tree. NMI = 1 would indicate complete agreement between species and OTU boundaries for all sequences analyzed. For this dataset, NMI peaks at OTU cutoff 0.08 and an NMI score of 0.98.
(DOC)
Genera as identified by RDP classifier, including distribution among individual samples.
(XLS)
Distribution of OTUs among individual samples after subsampling to 1227 sequences per sample, including OTU classification.
(XLS)
OTU overview, including OTU classification, aggregate information on OTU distribution among sample sets, and results of Metastats comparisons between sample sets (p< = 0.05).
(XLS)
List of Schaedler Flora sequences classified according to the criteria also employed for the experimental sequences.
(XLS)
Comparison of primer pairs 8F-338R and 8F-541R according to SILVA TestPrime results. Included groups correspond to the taxonomic identification of OTUs identified as restricted to one animal facility or significantly more abundant in one set of samples.
(XLS)
OTU classification according to (1) 80% consensus of classification of individual sequences and (2) classification of OTU consensus sequences (3) final OTU classification.
(XLS)
Funding Statement
This study was supported by grants DFG SFB 621/B8 and DFG SFB 900/Z1 from the German Research Foundation (DFG) to S.S., and National Institutes of Health (NIH) grants NIH P01-CA26731, NIH P30ES0026731 and NIH R01-OD011141 to J.G.F. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, et al. (1994) Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol 32: 1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ward JM, Fox JG, Anver MR, Haines DC, George CV, et al. (1994) Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J Natl Cancer Inst 86: 1222–1227. [DOI] [PubMed] [Google Scholar]
- 3. Fox JG, Yan L, Shames B, Campbell J, Murphy JC, et al. (1996) Persistent hepatitis and enterocolitis in germfree mice infected with Helicobacter hepaticus . Infect Immun 64: 3673–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boutin SR, Rogers AB, Shen Z, Fry RC, Love JA, et al. (2004) Hepatic temporal gene expression profiling in Helicobacter hepaticus-infected A/JCr mice. Toxicol Pathol 32: 678–693. [DOI] [PubMed] [Google Scholar]
- 5. Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, et al. (1997) Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus . Infect Immun 65: 3126–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kullberg MC, Ward JM, Gorelick PL, Caspar P, Hieny S, et al. (1998) Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect Immun 66: 5157–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Büchler G, Wos-Oxley ML, Smoczek A, Zschemisch N-H, Neumann D, et al. (2012) Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis 18: 943–954. [DOI] [PubMed] [Google Scholar]
- 8. Kullberg MC, Andersen JF, Gorelick PL, Caspar P, Suerbaum S, et al. (2003) Induction of colitis by a CD4+ T cell clone specific for a bacterial epitope. Proc Natl Acad Sci USA 100: 15830–15835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kullberg MC, Jankovic D, Gorelick PL, Caspar P, Letterio JJ, et al. (2002) Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J Exp Med 196: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maloy KJ, Salaun L, Cahill R, Dougan G, Saunders NJ, et al. (2003) CD4+CD25+ TR cells suppress innate immune pathology through cytokine-dependent mechanisms. J Exp Med 197: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nell S, Suerbaum S, Josenhans C (2010) The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol 8: 564–577. [DOI] [PubMed] [Google Scholar]
- 12. Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH (2011) Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol 4: 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, et al. (2012) Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA 109: E1820–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sterzenbach T, Bartonickova L, Behrens W, Brenneke B, Schulze J, et al. (2008) Role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J Bacteriol 190: 6398–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, et al. (2003) The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus . Proc Natl Acad Sci USA 100: 7901–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge Z, Rogers AB, Feng Y, Lee A, Xu S, et al. (2007) Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell Microbiol 9: 2070–2080. [DOI] [PubMed] [Google Scholar]
- 17. Bartonickova L, Sterzenbach T, Nell S, Kops F, Schulze J, et al. (2013) Hcp and VgrG1 are secreted components of the Helicobacter hepaticus type VI secretion system and VgrG1 increases the bacterial colitogenic potential. Cell Microbiol. 15: 992–1011. [DOI] [PubMed] [Google Scholar]
- 18. Chow J, Mazmanian SK (2010) A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7: 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boutin SR, Shen Z, Rogers AB, Feng Y, Ge Z, et al. (2005) Different Helicobacter hepaticus strains with variable genomic content induce various degrees of hepatitis. Infect Immun 73: 8449–8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ge Z, Sterzenbach T, Whary MT, Rickman BH, Rogers AB, et al. (2008) Helicobacter hepaticus HHGI1 is a pathogenicity island associated with typhlocolitis in B6.129-IL10tm1Cgn mice. Microbes Infect 10: 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sterzenbach T, Lee SK, Brenneke B, Von Goetz F, Schauer DB, et al. (2007) Inhibitory effect of enterohepatic Helicobacter hepaticus on innate immune responses of mouse intestinal epithelial cells. Infect Immun 75: 2717–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dieleman LA, Arends A, Tonkonogy SL, Goerres MS, Craft DW, et al. (2000) Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun 68: 5107–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whary MT, Taylor NS, Feng Y, Ge Z, Muthupalani S, et al. (2011) Lactobacillus reuteri promotes Helicobacter hepaticus-associated typhlocolitis in gnotobiotic B6.129P2-IL-10tm1Cgn (IL-10−/−) mice. Immunology 133: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, et al. (2010) Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS ONE 5: e8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, et al. (2007) Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell 131: 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Young VB, Knox KA, Pratt JS, Cortez JS, Mansfield LS, et al. (2004) In vitro and in vivo characterization of Helicobacter hepaticus cytolethal distending toxin mutants. Infect Immun 72: 2521–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pratt JS, Sachen KL, Wood HD, Eaton KA, Young VB (2006) Modulation of host immune responses by the cytolethal distending toxin of Helicobacter hepaticus . Infect Immun 74: 4496–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burich A, Hershberg R, Waggie K, Zeng W, Brabb T, et al. (2001) Helicobacter-induced inflammatory bowel disease in IL-10-and T cell-deficient mice. Am J Physiol Gastrointest Liver Physiol 281: G764–778. [DOI] [PubMed] [Google Scholar]
- 30. Schloss PD (2009) A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE 4: e8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ochman H, Worobey M, Kuo C-H, Ndjango J-BN, Peeters M, et al. (2010) Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, et al. (1999) Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol 65: 3287–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benson AK, Kelly SA, Legge R, Ma F, Low SJ, et al. (2010) Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107: 18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK (2010) Family relationship of female breeders reduce the systematic inter-individual variation in the gut microbiota of inbred laboratory mice. Lab Anim 44: 283–289. [DOI] [PubMed] [Google Scholar]
- 36. Ge Z, Feng Y, Taylor NS, Ohtani M, Polz MF, et al. (2006) Colonization dynamics of altered Schaedler flora is influenced by gender, aging, and Helicobacter hepaticus infection in the intestines of Swiss Webster mice. Appl Environ Microbiol 72: 5100–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vaahtovuo J, Toivanen P, Eerola E (2001) Study of murine faecal microflora by cellular fatty acid analysis; effect of age and mouse strain. Antonie Van Leeuwenhoek 80: 35–42. [DOI] [PubMed] [Google Scholar]
- 38. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. The Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peña JA, Rogers AB, Ge Z, Ng V, Li SY, et al. (2005) Probiotic Lactobacillus spp. diminish Helicobacter hepaticus-induced inflammatory bowel disease in interleukin-10-deficient mice. Infect Immun 73: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazmanian SK, Round JL, Kasper DL (2008) A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453: 620–625. [DOI] [PubMed] [Google Scholar]
- 42. Rajilić-Stojanović M, Biagi E, Heilig HGHJ, Kajander K, Kekkonen RA, et al. (2011) Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141: 1792–1801. [DOI] [PubMed] [Google Scholar]
- 43. Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, et al. (2012) Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol 13: R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, et al. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kulagina EV, Efimov BA, Maximov PY, Kafarskaia LI, Chaplin AV, et al. (2012) Species composition of Bacteroidales order bacteria in the feces of healthy people of various ages. Biosci Biotechnol Biochem 76: 169–171. [DOI] [PubMed] [Google Scholar]
- 46. Kuehbacher T, Rehman A, Lepage P, Hellmig S, Fölsch UR, et al. (2008) Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol 57: 1569–1576. [DOI] [PubMed] [Google Scholar]
- 47. Jia W, Whitehead RN, Griffiths L, Dawson C, Bai H, et al. (2012) Diversity and distribution of sulphate-reducing bacteria in human faeces from healthy subjects and patients with inflammatory bowel disease. FEMS Immunol Med Microbiol 65: 55–68. [DOI] [PubMed] [Google Scholar]
- 48. Baron EJ, Summanen P, Downes J, Roberts MC, Wexler H, et al. (1989) Bilophila wadsworthia, gen. nov. and sp. nov., a unique gram-negative anaerobic rod recovered from appendicitis specimens and human faeces. J Gen Microbiol 135: 3405–3411. [DOI] [PubMed] [Google Scholar]
- 49. Finegold S, Summanen P, Hunt Gerardo S, Baron E (1992) Clinical importance of Bilophila wadsworthia . Eur J Clin Microbiol Infect Dis 11: 1058–1063. [DOI] [PubMed] [Google Scholar]
- 50. Schumacher UK, Eiring P, Häcker F-M (1997) Incidence of Bilophila wadsworthia in appendiceal, peritoneal and fecal samples from children. Clin Microbiol Infect 3: 134–136. [DOI] [PubMed] [Google Scholar]
- 51. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, et al. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ciszek-Lenda M, Strus M, Gorska-Fraczek S, Targosz-Korecka M, Srottek M, et al. (2011) Strain specific immunostimulatory potential of lactobacilli-derived exopolysaccharides. Central Eur J Immunol 36: 121–129. [Google Scholar]
- 53. Liu Y, Fatheree NY, Mangalat N, Rhoads JM (2010) Human-derived probiotic Lactobacillus reuteri strains differentially reduce intestinal inflammation. Am J Physiol Gastrointest Liver Physiol 299: G1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Azevedo MSP, Zhang W, Wen K, Gonzalez AM, Saif LJ, et al. (2012) Lactobacillus acidophilus and Lactobacillus reuteri modulate cytokine responses in gnotobiotic pigs infected with human rotavirus. Benef Microbes 3: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hoffmann M, Rath E, Hölzlwimmer G, Quintanilla-Martinez L, Loach D, et al. (2008) Lactobacillus reuteri 100–23 transiently activates intestinal epithelial cells of mice that have a complex microbiota during early stages of colonization. J Nutr 138: 1684–1691. [DOI] [PubMed] [Google Scholar]
- 56.Wedekind D, Reifenberg K, Hedrich HJ (2012) Genetic monitoring of inbred strains of mice. In: Hedrich HJ, editor. The laboratory mouse. Amsterdam: Elsevier. 621–637.
- 57. Nicklas W, Baneux P, Boot R, Decelle T, Deeny AA, et al. (2002) Recommendations for the health monitoring of rodent and rabbit colonies in breeding and experimental units. Lab Anim 36: 20–42. [DOI] [PubMed] [Google Scholar]
- 58. Ge Z, Feng Y, Whary MT, Nambiar PR, Xu S, et al. (2005) Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect Immun 73: 3559–3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ge Z, White DA, Whary MT, Fox JG (2001) Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus. J Clin Microbiol 39: 2598–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, et al. (2009) A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lofgren JL, Whary MT, Ge Z, Muthupalani S, Taylor NS, et al. (2011) Lack of commensal flora in Helicobacter pylori–infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 140: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, et al. (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schloss PD (2011) Schloss SOP - mothur. Available: http://www.mothur.org/wiki/Schloss_SOP. Accessed 2011 Oct 28.
- 64. Schloss PD, Gevers D, Westcott SL (2011) Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schloss PD (2011) Silva reference files - mothur. Available: http://www.mothur.org/wiki/Silva_reference_files. Accessed 2011 Oct 31.
- 66. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, et al. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Giongo A, Davis-Richardson AG, Crabb DB, Triplett EW (2010) TaxCollector: Modifying current 16S rRNA databases for the rapid classification at six taxonomic levels. Diversity 2: 1015–1025. [Google Scholar]
- 69. Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214. [DOI] [PubMed] [Google Scholar]
- 70. Cai Y, Sun Y (2011) ESPRIT-Tree: hierarchical clustering analysis of millions of 16S rRNA pyrosequences in quasilinear computational time. Nucleic Acids Res 39: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fred ALN, Jain AK (2003) Robust data clustering. Proceedings IEEE Conference on Computer Vision and Pattern Recognition. 128–136.
- 72.Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, et al. (2012) Geneious. Available: http://www.geneious.com.
- 73. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nahid A, Bowne J, Murray G (2011) heatmap. Metabolomics Australia, Bioinformatics group. Available: http://code.google.com/p/ma-bioinformatics/downloads/list. Accessed 2011 Dec20.
- 76. White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Normalized mutual information (NMI) value between species and OTUs obtained for 30 difference levels using ESPRIT-Tree. NMI = 1 would indicate complete agreement between species and OTU boundaries for all sequences analyzed. For this dataset, NMI peaks at OTU cutoff 0.08 and an NMI score of 0.98.
(DOC)
Genera as identified by RDP classifier, including distribution among individual samples.
(XLS)
Distribution of OTUs among individual samples after subsampling to 1227 sequences per sample, including OTU classification.
(XLS)
OTU overview, including OTU classification, aggregate information on OTU distribution among sample sets, and results of Metastats comparisons between sample sets (p< = 0.05).
(XLS)
List of Schaedler Flora sequences classified according to the criteria also employed for the experimental sequences.
(XLS)
Comparison of primer pairs 8F-338R and 8F-541R according to SILVA TestPrime results. Included groups correspond to the taxonomic identification of OTUs identified as restricted to one animal facility or significantly more abundant in one set of samples.
(XLS)
OTU classification according to (1) 80% consensus of classification of individual sequences and (2) classification of OTU consensus sequences (3) final OTU classification.
(XLS)