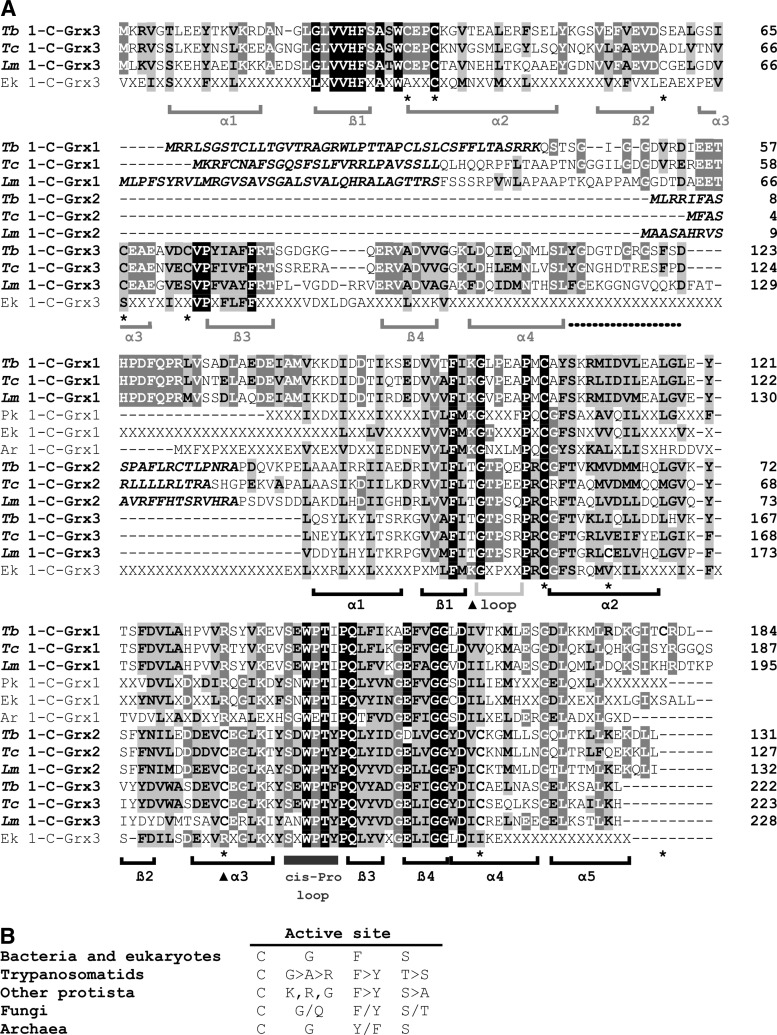
FIG. 2.
Sequence analysis of monothiol glutaredoxins. (A) 1-C-Grxs from trypanosomatids (Tb, Trypanosoma brucei; Tc, Trypanosoma cruzi; Lm, Leishmania major) were aligned with the consensus sequences obtained for prokaryote (Pk), eukaryote (Ek), and archeabacteria (Ar) homologues using the ClustalW algorithm (85) and manually adjusted as necessary. Accession numbers are AJ619696, AM489503 and AM489504 for T. brucei 1-C-Grx1, 2 and 3, respectively; XP_807837, XP_803206 and XP_813048 for T. cruzi 1-C-Grx1, 2 and 3, respectively; NP_047037, CAJ01951 and XP_843232 for L. major 1-C-Grx1, 2 and 3, respectively. Residues given in italic indicate the predicted mitochondrial targeting sequence of Kinetoplastida 1-C-Grx1 and 2. Residues shown in black on gray and white on gray represent residues that are similar and identical, respectively, in at least 40% of the aligned sequences. Residues that are strictly conserved in all sequences analyzed are shown white on black. Cysteine residues in the 1-C-Grxs from trypanosomatids are indicated with an asterisk at the bottom of the alignment. The arrow heads mark basic residues suggested to be involved in glutathione binding in classical Grxs (28, 40). The consensus sequences for the three phylogenetic domains were obtained by alignment of characterized or putative 1-C-Grxs from representative organisms. The sequences used for prokaryotes were from Agrobacterium tumefaciens (α-protobacterium, Acc. Nr. AAK87621.1), Neisseria gonorrheae (β-protobacterium, YP_207507.1), Myxococcus xanthus (δ-protobacterium, ABF89434.1), Escherichia coli (γ-protobacterium, 1YKA) and Synechococcus elongatus (cyanobacterium, BAD78595.1); for archaeabacteria: Haloarcula marismortui (AAV46243.1), Natronomonas pharaonis (YP_326686.1), Haloquadratum walsbyi (CAJ51793.1), and Halobacterium sp. (AAG18993.1); for eukaryotes: Gallus gallus (NP_001008472.1 and XP_421826.1), Danio rerio (AAH59659.1 and NP_001005950.1), Homo sapiens (NP_057501.2 and AAH05289.1), Caenorhabditis elegans (CAB11547.1 and NP_001023756.1), Apis mellifera (XP_625213.1 and XP_392870.1), Tetraodon negroviridis (CAG00128.1 and CAG02746.1), Saccharomyces cerevisiae (Q02784, Q03835 and P32642), Arabidopsis thaliana (AY157988), Tribolium castaneum (XP_975383.1 and XP_972466.1), Porphyra purpurea (P51384), Bos taurus (XP_582303.1 and AAX46537.1), and Xenopus tropicalis (AAH75374.1 and NP_001017209.1). The consensus sequences show only residues that were common to more than 75% of the proteins analyzed. X represents any amino acid. The secondary structure motifs below the alignment refer to the α-helical (α) and ß-sheet (ß) regions in the NMR structure of T. brucei 1-C-Grx1 (black) as well as the homology model of the Trx domain of T. brucei 1-C-Grx3 (gray). The dotted line marks the linker region of T. brucei 1-C-Grx3 and the light and dark gray bar indicates the insertion preceding the active site and the cys-Pro loop. (B) The consensus active site motif of 1-C-Grxs from representatives of the prokaryotic and eukaryotic domains and the family of Trypanosomatidae was obtained from the sequences listed above. The following sequences were used for other protista: Cryptosporidium parvum (CAD98438), Tetrahymena thermophila (XP 001016225, XP 001032143 and XP 001008985), Paramecium tetraurelia (CAK57552, CAK90692 and CAK55785), and Plasmodium falciparum (CAG25239 and CAD50844, for Glp2 and Glp3, respectively); fungi: Encephalitozoon cuniculi (NP 597481), Cocidioides immitis (XP 001244791), and Mortierella alpina (CAB 56513); Archaea: Haloarcula morismortui (AAV46243), Natromonas pharaonis (YP326686), and Haloquadratum walsbyi (CAJ 51793).