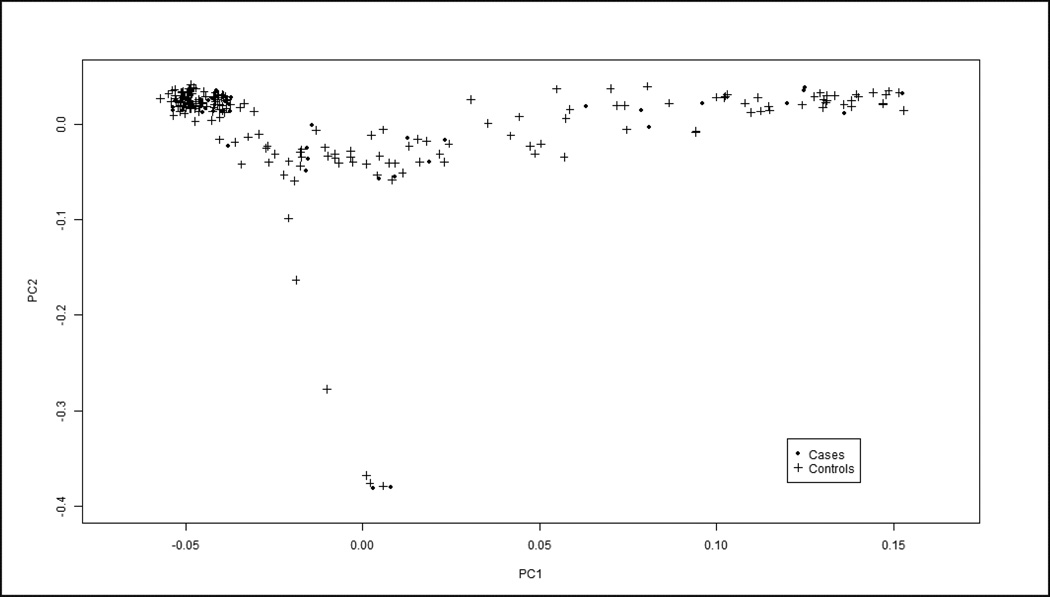
Figure 1.
Principal Component Analysis (PCA) was performed to extrapolate ethnicity based on genotype information of our study population. Although our two HCV datasets were ethnically heterogeneous, both IIT cases (dots) and HCV controls (crosses) showed similar distributions and while they did not cluster at any one corner of the graph (as would happen in an ethnically homogenous sample), their compatibility enabled us to compare them in our association analyses. Moreover, the PCA showed that the lambda score, or genomic inflation factor, was 1 which implies that the data is not confounded by effects of population stratification.