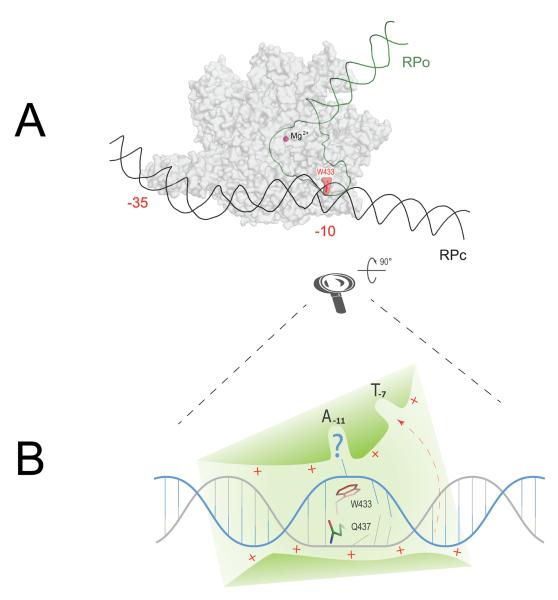
Figure 2.
Structural modeling of −10 element recognition as the first step in promoter opening. (A) Schematic comparison of closed (RPc) and open (RPo) promoter complexes and suggested wedge role for W433 as an initiator of promoter melting. RNAP shown as a gray semi-transparent surface. DNA in RPc is shown black, downstream portion of promoter DNA after the melting (RPo) is green. The wedge W433 is highlighted in red. Catalytic Mg2+ = purple sphere. −35 and −10 promoter elements are labeled. (B) Schematic close-up of the first step in −10 element recognition. DNA directed in the shallow trough on RNAP surface (green) via a steric and electrostatic fit. Non-template strand is shown in blue, template strand is gray. The helix invasion by W433 disrupts one of the base pairs and flips out the non-template base into A-11 pocket, thereby making the upstream neighboring template-strand base accessible for H-bonding with Q437 (this amino acid residue was implicated in −12 base pair recognition in genetic screens.58,59 Following the recognition of the T–12A–11 step, DNA untwisting will proceed downstream, accompanied by T–7 flipping out into the respective σ-pocket (shown by red dashed arrow).