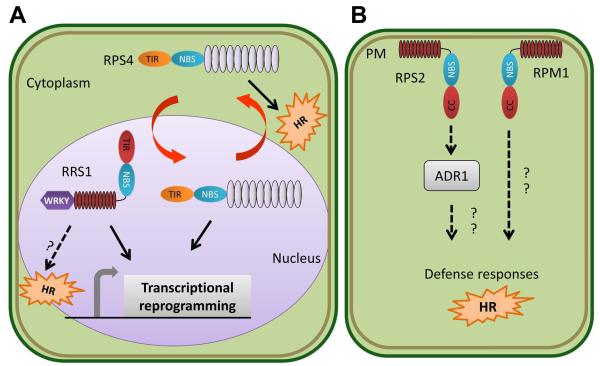
Fig. 2.
Subcellular partitioning of NLR-mediated plant immune responses. (a) RPS4 and RRS1 mediate activation of plant immune responses against Pseudomonas and Ralstonia bacterial pathogens carrying the cognate effectors AvrRps4 and PopP2. RPS4 is a TIR-NLR which predominately localizes to endomembranes in a resting state and upon activation undergoes shuttling between the cytoplasm and nucleus. Nuclear localization is required for RPS4-mediated transcriptional reprogramming, while recognition in the cytoplasm is required for the HR. However, however robust HR and sustained transcriptional reprogramming requires coordination between nuclear and cytoplasmic pools of activated RPS4. RRS1 exhibits novel domain architecture, with a TIR-NLR fused to a C-terminal WRKY domain. WRKY domains are found in multiple plant transcription factors required for innate immune reprogramming. RRS1 is nuclear localized, presumably where its WRKY domain acts as a transcriptional regulator upon recognition of PopP2, leading to the transcription of defense related genes. (b) RPS2 and RPM1, two CC-NLRs that recognize Pseudomonas effectors AvrRpt2, and AvrRpm1/ AvrB respectively, reside at the plasma membrane. RPM1’s subcellular localization does not change in response to activation. Thus, dynamic re-localization of plant immune receptors between cellular compartments is not necessary for all NLRs. RPS2 downstream signaling also requires the ‘helper’ NLR ADR1, which may interface with components relaying the activation of defense responses in other subcellular compartments.